
The evaluation of the matching results for the anatomy data set is problematic as many standard ways of doing an evaluation do cannot be applied in this case. First of all it is not possible to compare generated mappings against a gold standard mapping, because such a mapping does not exist for these particular ontologies and creating one is not feasible due to the size of the ontologies and the expertise necessary. The same holds for a manual evaluation of the generated mappings as the four systems participating in this task together produced more than 8000 mappings. Previous plans for automatically generating a subset of the gold standard mapping similar to the evaluation of the directory task turned out to fail on this data set due to the special nature of the ontologies.
In the face of these difficulties we decided to perform a weaker form of evaluation that does not really measure the precision and recall of the different systems but analyzes commonalities and differences of the results of the different systems as well as the coverage of the terminology in the ontologies. In particular, we looked at the following aspects of the mapping sets generated by the participating tools:
This means that instead of directly measuring precision and recall we concentrate on indicators that gives hints towards the probable precision and recall of a system. The coverage of the terminology is a (partial) indicator for the recall of the matching system. The percentage of mappings that have also been found by other systems is a (partial) indicator for the precision as mappings found by different systems are more likely to be correct than mappings only found by a single system. It is important to note that these indicators do not provide a sufficient basis for comparing system at a quantitative level. All we can provide are tendencies.
The evaluation showed that the four systems that participated in the task together produced mappings for between 2000 and 3000 terms in GALEN. The COMA++ system is an exception as it only identified mappings for about 600 concepts. We will comment on this later one.
| Terms mapped | |
| NIH | 2966 |
| Islab | 2963 |
| PRIOR | 2590 |
| falcon | 2204 |
| COMA++ | 622 |
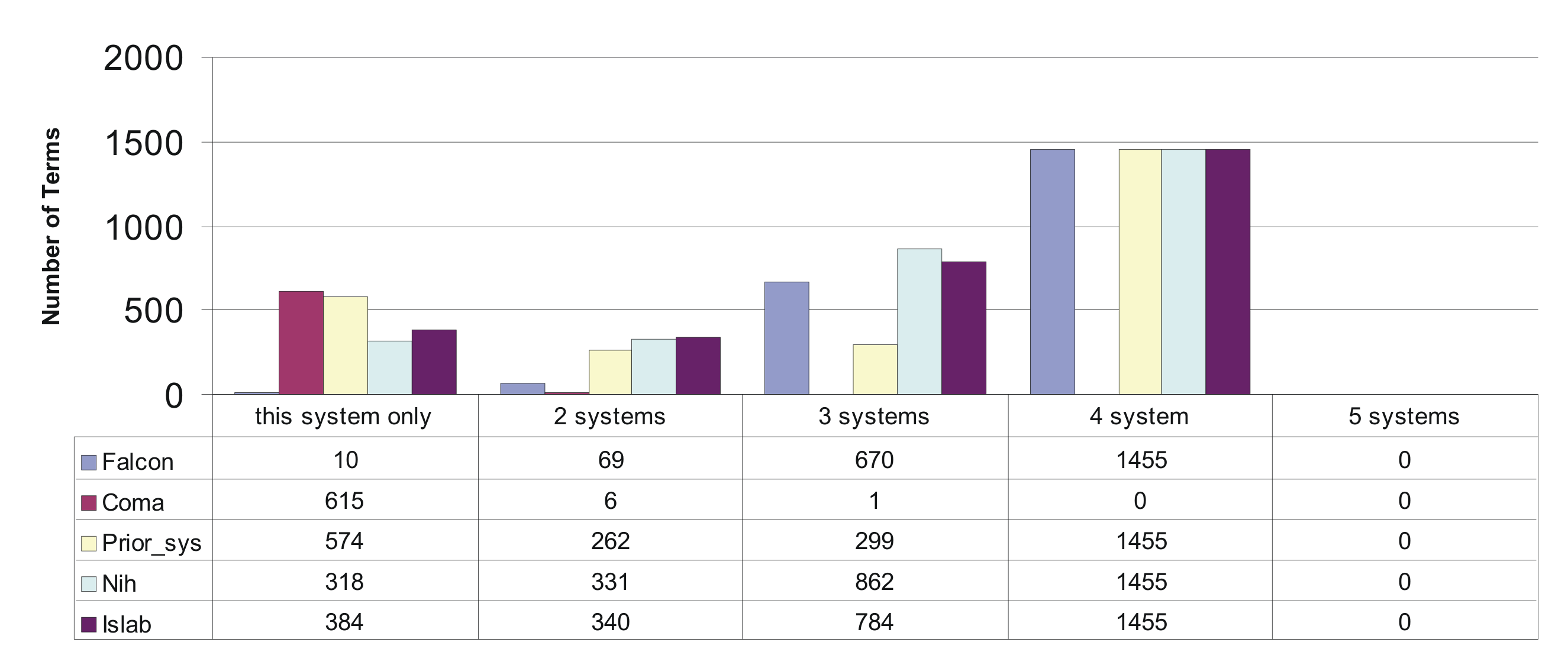
The percentages given above show that there is a significant overlap in the sets of terms that could be mapped by the different systems. This is expected as obvious matches should be found by all systems. Having a closer look at the overlap between the terms covered, revealed that none of the terms have been mapped by allsystems. About 1500 terms were mapped by 4 of the 5 systems. Surprisingly we observed a significant overlap between NIH, ISlab, PRIOR and falcon, whereas COMA produced a mappings for a set of terms almost completely disjoint from the other ones. Only 6 of the 622 Concepts covered by COMA where also found by other systems. The following figure provides the exact figures.

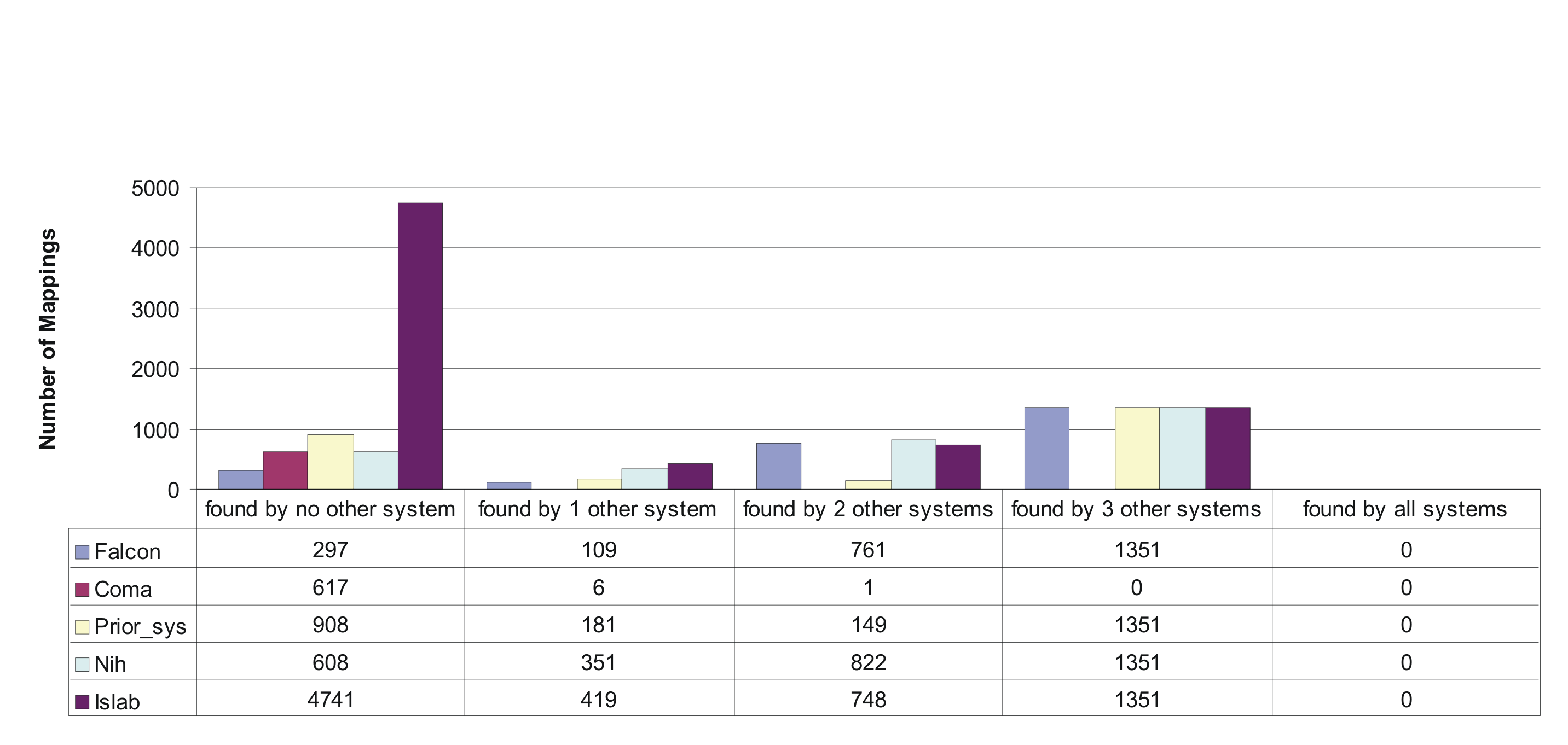
When looking at the numbers in the figure, we see that falcon is the system with the highest degree of overlap with other systems. NIH also has a high number of mappings that have been found by other systems as well, but it also has a significant amount of mappings not found by any other system. For the prior system we observe an even larger number of mappings not found by any other system.
As already mentioned previously, the evaluation carried out does not allow us to really judge the quality of the matching systems of to rank them in any way. The evaluation, however provides some insights in the trade-off systems make with respect to precision and recall and shed some light on typical problems of current matching systems.
The first conclusion is that the results confirm the observation that the falcon system seems to emphasize correctness of matching results. This is documented in a relatively low number of mappings moist of which have also been found by at least one other system. The ISlab system on the other hand seems to focus more on recall. This is documented in a high number of mappings found as well as in the fact that the ISlab results contain a very high number of mappings that have also been found by at least one other system.
The results of the COMA system deserve special attention. As we see, the results of COMA are almost disjoint with the result of the other systems. We have analyzed this phenomenon in more detail. It appears that this phenomenon is caused by irregularities in class names occurring in the GALEN ontology. Besides regular class names like Cell there also is a significant number of concepts with compositional names (e.g. _--ControlOfFlow_which___hasEffectiveness_--Effectiveness_which___hasAbsoluteState_effective__--__--). When looking at the results of the different systems it turns out that none of the systems except for COMA was unable to map these compositional names with the effect that corresponding mappings are not present in the results. The results of COMA on the other hand managed to compute mappings for these concept, but surprisingly did not produce any mapping for simple class names. The result is a mapping set that is almost disjoint from the other results. This also means that it is hard to say anything about the results produced by COMA expect that they cover a part of the GALEN ontology that is ignored by other systems.
For future editions we think that the anatomy data set is still challenging for existing matching systems which is clearly documented by the fact that none of the matchers was able to cover all of the concepts in GALEN. Ideally, the experiments will be repeated and an evaluation will be conducted based on a manual inspection of the mapping sets produced by this years participants in order to be able to make more precise assertions about the accuracy of matching results.
For the first time since the anatomy data set has been used in the ontology alignment evaluation challenge, we are in a position, were we were actually able to compare the results of different matching systems. The results show that there is still a lot of work to do to make matching systems ready for real life applications. The problems above showed that differences in the naming scheme of classes can already cause matchers to fail on a significant subset of the vocabulary (note that this holds for all the systems in the evaluation, the COMA++ system just worked on the smaller subset which affects the results). For us this is a clear sign that this benchmark is still a good one that should by included in following challenges as well.
In order to enable people to improve their systems based on this data set, it is important to provide a more informative evaluation next time. The data we have now provides the basis for generating a partial gold standard mapping that can be used in the next evaluation campaign. In particular, the set of mappings found by 4 of the 5 systems independently is a good starting point for a quantitative evaluation. This set contains about 1400 mappings which can be evaluated manually to form the core of a gold standard mapping. Based on this gold standard mapping, we can estimate the recall of matching systems in the same way done for the directory mapping set. Such a gold standard mapping also eases the estimation of the precision of mappings, as only those mappings not in the golden standard have to be checked manually. The results of such a manual process will again feed into the gold standard making it more and more complete over time.