
Like last year, 13 systems participated in the benchmark track of this year's campaign.
The evaluation has been performed on the files provided by the participants (available here) which have been processed by the following script). The full set of normalised alignments is also available.
Table 1 provides the consolidated results by groups of tests. We display the results of participants as well as those given by some simple edit distance algorithm on labels (edna). The computed values are real precision and recall and not an average of precision and recall.
These results show already that the three systems which last year were ahead of the pack, are still at the top with are relatively ahead (ASMOV, Lily and RiMOM) with three close followers (AROMA, DSSim, and Anchor-Flood replacing Falcon, Prior+ and OLA2 last year). No system had strictly lower performance than edna. Each algorithm has its best score with the 1xx test series. There is no particular order between the two other series.
| algo | refalign | edna | aflood | aroma | ASMOV | CIDER | DSSim | GeRoMe | Lily | MapPSO | RiMOM | SAMBO | SAMBOdtf | SPIDER | TaxoMap | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. |
| 1xx | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.96 | 0.79 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 0.34 |
| 2xx | 1.00 | 1.00 | 0.41 | 0.56 | 0.96 | 0.69 | 0.96 | 0.70 | 0.95 | 0.85 | 0.97 | 0.57 | 0.97 | 0.64 | 0.56 | 0.52 | 0.97 | 0.86 | 0.48 | 0.53 | 0.96 | 0.82 | 0.98 | 0.54 | 0.98 | 0.56 | 0.97 | 0.57 | 0.95 | 0.21 |
| 3xx | 1.00 | 1.00 | 0.47 | 0.82 | 0.95 | 0.66 | 0.82 | 0.71 | 0.81 | 0.77 | 0.90 | 0.75 | 0.90 | 0.71 | 0.61 | 0.40 | 0.87 | 0.81 | 0.49 | 0.25 | 0.80 | 0.81 | 0.95 | 0.80 | 0.91 | 0.81 | 0.15 | 0.81 | 0.92 | 0.21 |
| H-mean | 1.00 | 1.00 | 0.43 | 0.59 | 0.97 | 0.71 | 0.95 | 0.70 | 0.95 | 0.86 | 0.97 | 0.62 | 0.97 | 0.67 | 0.60 | 0.58 | 0.97 | 0.88 | 0.51 | 0.54 | 0.96 | 0.84 | 0.99 | 0.58 | 0.98 | 0.59 | 0.81 | 0.63 | 0.91 | 0.22 |
After remarks of last year we made two changes on the tests this year:
| algo | refalign | edna | aflood | aroma | ASMOV | CIDER | DSSim | GeRoMe | Lily | MapPSO | RiMOM | SAMBO | SAMBOdtf | SPIDER | TaxoMap | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. |
| 1xx | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.96 | 0.79 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 0.34 |
| 2xx | 1.00 | 1.00 | 0.41 | 0.56 | 0.96 | 0.69 | 0.96 | 0.70 | 0.95 | 0.85 | 0.97 | 0.57 | 0.97 | 0.64 | 0.56 | 0.52 | 0.97 | 0.86 | 0.48 | 0.53 | 0.96 | 0.82 | 0.98 | 0.54 | 0.98 | 0.56 | 0.97 | 0.57 | 0.95 | 0.21 |
| 3xx | 1.00 | 1.00 | 0.47 | 0.82 | 0.95 | 0.66 | 0.82 | 0.71 | 0.81 | 0.77 | 0.90 | 0.75 | 0.90 | 0.71 | 0.61 | 0.40 | 0.87 | 0.81 | 0.49 | 0.25 | 0.80 | 0.81 | 0.95 | 0.80 | 0.91 | 0.81 | 0.15 | 0.81 | 0.92 | 0.21 |
| H-mean | 1.00 | 1.00 | 0.45 | 0.61 | 0.97 | 0.71 | 0.96 | 0.72 | 0.95 | 0.85 | 0.97 | 0.62 | 0.97 | 0.68 | 0.59 | 0.54 | 0.96 | 0.87 | 0.52 | 0.55 | 0.95 | 0.83 | 0.98 | 0.59 | 0.98 | 0.61 | 0.67 | 0.62 | 0.95 | 0.22 |
Table 3 provides results on the very easy tests of 2004. As expected, systems perform very well.
| algo | refalign | edna | aflood | aroma | ASMOV | CIDER | DSSim | GeRoMe | Lily | MapPSO | RiMOM | SAMBO | SAMBOdtf | SPIDER | TaxoMap | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. |
| 1xx | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.96 | 0.79 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 0.34 |
| 2xx | 1.00 | 1.00 | 0.67 | 0.74 | 0.98 | 0.92 | 0.99 | 0.94 | 0.99 | 0.98 | 0.98 | 0.82 | 0.98 | 0.89 | 0.93 | 0.72 | 0.99 | 0.98 | 0.66 | 0.72 | 1.00 | 0.98 | 0.99 | 0.75 | 0.99 | 0.77 | 0.98 | 0.82 | 0.94 | 0.24 |
| 3xx | 1.00 | 1.00 | 0.47 | 0.82 | 0.95 | 0.66 | 0.82 | 0.71 | 0.81 | 0.77 | 0.90 | 0.75 | 0.90 | 0.71 | 0.61 | 0.40 | 0.87 | 0.81 | 0.49 | 0.25 | 0.80 | 0.81 | 0.95 | 0.80 | 0.91 | 0.81 | 0.15 | 0.81 | 0.92 | 0.21 |
| H-mean | 1.00 | 1.00 | 0.67 | 0.80 | 0.98 | 0.90 | 0.97 | 0.92 | 0.97 | 0.96 | 0.97 | 0.84 | 0.97 | 0.88 | 0.90 | 0.69 | 0.98 | 0.96 | 0.70 | 0.71 | 0.97 | 0.96 | 0.98 | 0.80 | 0.98 | 0.81 | 0.57 | 0.85 | 0.95 | 0.25 |
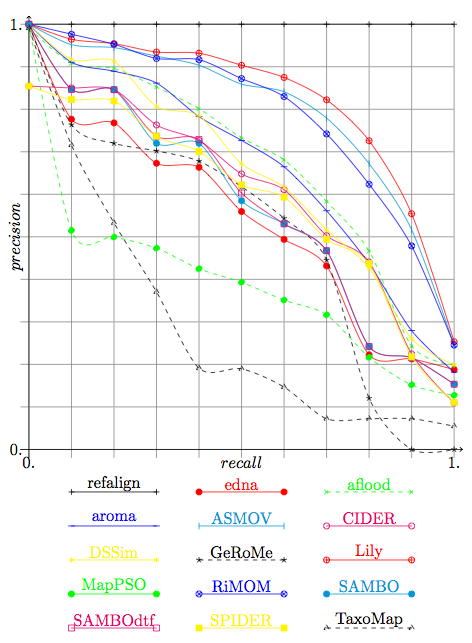
This year again the apparently best algorithms provided their results with confidence measures. It is thus possible to draw precision/recall graphs in order to compare them.
We provide in Figure 1 the precision and recall graphs of this year. They are only relevant for the results of participants who provided confidence measures different from 1 or 0 (see the table in the Summary page). This graph has been drawn with only technical adaptation of the technique used in TREC. Moreover, due to lack of time, these graphs have been computed by averaging the graphs of each of the tests (instead to pure precision and recall). They do not feature the curves of previous years since the test sets have been changed.
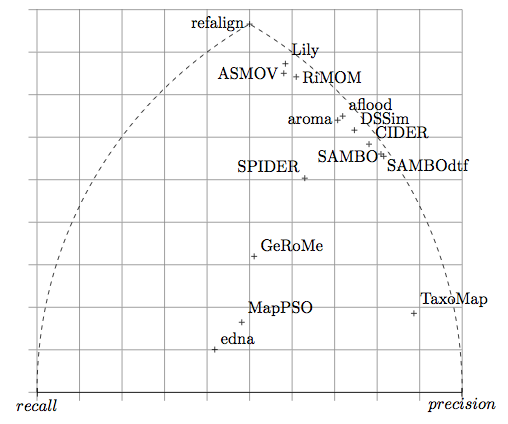
These results and those displayed in Figure 2 single out a group of systems, ASMOV, Lily, and RiMOM which seem to perform these tests at the highest level of quality. So this confirm the lead that we see on raw results.
Like the two previous years, there is a gap between these systems and their followers. The gap between these systems and the next ones (AROMA, DSSim, and Anchor-Flood) has reformed. It was filled last year by Falcon and OLA and Prior which did not participate this year.
The triangular display shows the same results as the other representations.
| algo | refalign | edna | aflood | aroma | ASMOV | CIDER | DSSim | GeRoMe | Lily | MapPSO | RiMOM | SAMBO | SAMBOdtf | SPIDER | TaxoMap | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. | Prec. | Rec. |
| 101 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.34 |
| 103 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 1.00 | 0.94 | 0.79 | 1.00 | 1.00 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 0.34 |
| 104 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.95 | 0.79 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.34 |
| 201 | 1.00 | 1.00 | 0.04 | 0.04 | 0.96 | 0.84 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.60 | 0.95 | 0.94 | 0.87 | 0.79 | 1.00 | 1.00 | 0.12 | 0.13 | 1.00 | 1.00 | 0.92 | 0.25 | 0.92 | 0.25 | 1.00 | 0.60 | 0.75 | 0.06 |
| 201-2 | 1.00 | 1.00 | 0.76 | 0.79 | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.90 | 0.99 | 0.99 | 0.95 | 0.79 | 1.00 | 1.00 | 0.79 | 0.88 | 1.00 | 1.00 | 1.00 | 0.84 | 1.00 | 0.84 | 0.98 | 0.90 | 0.97 | 0.30 |
| 201-4 | 1.00 | 1.00 | 0.57 | 0.60 | 1.00 | 0.79 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.87 | 0.98 | 0.98 | 0.93 | 0.79 | 1.00 | 1.00 | 0.66 | 0.70 | 1.00 | 1.00 | 0.97 | 0.70 | 0.97 | 0.70 | 1.00 | 0.87 | 0.92 | 0.25 |
| 201-6 | 1.00 | 1.00 | 0.40 | 0.41 | 0.98 | 0.91 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.82 | 0.96 | 0.95 | 0.97 | 0.79 | 1.00 | 1.00 | 0.50 | 0.56 | 1.00 | 1.00 | 0.97 | 0.58 | 0.97 | 0.58 | 1.00 | 0.82 | 0.90 | 0.20 |
| 201-8 | 1.00 | 1.00 | 0.23 | 0.24 | 0.95 | 0.58 | 1.00 | 0.99 | 1.00 | 1.00 | 0.93 | 0.69 | 0.95 | 0.94 | 0.91 | 0.79 | 1.00 | 1.00 | 0.28 | 0.31 | 1.00 | 1.00 | 0.95 | 0.41 | 0.95 | 0.41 | 0.93 | 0.69 | 0.86 | 0.12 |
| 202 | 1.00 | 1.00 | 0.06 | 0.06 | 1.00 | 0.78 | 0.95 | 0.40 | 0.92 | 0.80 | n/a | n/a | 1.00 | 0.24 | 0.17 | 0.06 | 1.00 | 0.84 | 0.05 | 0.05 | 1.00 | 0.81 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 202-2 | 1.00 | 1.00 | 0.76 | 0.79 | 1.00 | 0.86 | 0.99 | 0.88 | 0.99 | 0.97 | 1.00 | 0.82 | 0.98 | 0.82 | 0.74 | 0.77 | 1.00 | 0.97 | 0.72 | 0.81 | 1.00 | 0.97 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.82 | 0.84 | 0.22 |
| 202-4 | 1.00 | 1.00 | 0.57 | 0.60 | 1.00 | 0.92 | 0.97 | 0.78 | 0.96 | 0.93 | 1.00 | 0.66 | 0.99 | 0.69 | 0.93 | 0.67 | 1.00 | 0.92 | 0.55 | 0.60 | 1.00 | 0.93 | 1.00 | 0.60 | 1.00 | 0.60 | 1.00 | 0.66 | 0.89 | 0.18 |
| 202-6 | 1.00 | 1.00 | 0.40 | 0.41 | 1.00 | 0.88 | 0.99 | 0.68 | 0.92 | 0.88 | 0.96 | 0.46 | 0.98 | 0.56 | 0.94 | 0.60 | 0.98 | 0.87 | 0.34 | 0.37 | 1.00 | 0.88 | 1.00 | 0.41 | 1.00 | 0.41 | 0.96 | 0.46 | 0.85 | 0.11 |
| 202-8 | 1.00 | 1.00 | 0.23 | 0.24 | 0.91 | 0.42 | 0.96 | 0.56 | 0.93 | 0.85 | 1.00 | 0.27 | 0.95 | 0.40 | 0.77 | 0.48 | 0.98 | 0.85 | 0.20 | 0.23 | 1.00 | 0.87 | 1.00 | 0.22 | 1.00 | 0.22 | 1.00 | 0.27 | 0.83 | 0.05 |
| 203 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.95 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.81 | 0.26 |
| 204 | 1.00 | 1.00 | 0.90 | 0.94 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.97 | 0.99 | 0.78 | 1.00 | 0.79 | 1.00 | 1.00 | 0.85 | 0.93 | 1.00 | 1.00 | 1.00 | 0.94 | 1.00 | 0.96 | 0.98 | 0.97 | 0.87 | 0.27 |
| 205 | 1.00 | 1.00 | 0.34 | 0.35 | 0.89 | 0.57 | 1.00 | 0.99 | 1.00 | 0.99 | 0.96 | 0.76 | 0.89 | 0.80 | 1.00 | 0.79 | 1.00 | 0.99 | 0.30 | 0.33 | 1.00 | 0.99 | 0.93 | 0.41 | 0.92 | 0.46 | 0.96 | 0.76 | 0.64 | 0.09 |
| 206 | 1.00 | 1.00 | 0.50 | 0.53 | 0.96 | 0.84 | 1.00 | 0.98 | 1.00 | 0.99 | 0.95 | 0.64 | 0.95 | 0.90 | 0.93 | 0.78 | 1.00 | 0.99 | 0.35 | 0.38 | 1.00 | 0.99 | 1.00 | 0.44 | 1.00 | 0.55 | 0.95 | 0.64 | NaN | 0.00 |
| 207 | 1.00 | 1.00 | 0.50 | 0.53 | 0.96 | 0.84 | 1.00 | 0.98 | 1.00 | 0.99 | 0.92 | 0.62 | 0.96 | 0.91 | 0.93 | 0.78 | 1.00 | 0.99 | 0.35 | 0.39 | 1.00 | 0.99 | 1.00 | 0.44 | 1.00 | 0.55 | 0.92 | 0.62 | NaN | 0.00 |
| 208 | 1.00 | 1.00 | 0.90 | 0.94 | 1.00 | 0.96 | 1.00 | 0.93 | 1.00 | 1.00 | 1.00 | 0.98 | 0.97 | 0.72 | 0.99 | 0.77 | 1.00 | 0.99 | 0.78 | 0.88 | 1.00 | 1.00 | 1.00 | 0.88 | 1.00 | 0.90 | 1.00 | 0.98 | 0.76 | 0.23 |
| 209 | 1.00 | 1.00 | 0.33 | 0.34 | 0.91 | 0.49 | 0.84 | 0.61 | 0.91 | 0.88 | 0.84 | 0.27 | 0.85 | 0.40 | 0.58 | 0.45 | 0.97 | 0.88 | 0.22 | 0.25 | 0.87 | 0.86 | 0.86 | 0.26 | 0.82 | 0.33 | 0.84 | 0.27 | 0.43 | 0.03 |
| 210 | 1.00 | 1.00 | 0.51 | 0.54 | 0.96 | 0.80 | 0.92 | 0.69 | 0.96 | 0.91 | 1.00 | 0.33 | 0.97 | 0.39 | 0.61 | 0.73 | 1.00 | 0.89 | 0.18 | 0.20 | 0.96 | 0.89 | 1.00 | 0.23 | 1.00 | 0.42 | 1.00 | 0.33 | 0.67 | 0.04 |
| 221 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.65 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.34 |
| 222 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.97 | 0.78 | 1.00 | 1.00 | 0.91 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 0.97 | 0.31 |
| 223 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 0.95 | 0.95 | 0.99 | 0.99 | 0.97 | 0.97 | 1.00 | 1.00 | 0.85 | 0.78 | 0.98 | 0.98 | 0.96 | 0.89 | 1.00 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 | 0.97 | 0.97 | 0.97 | 0.31 |
| 224 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.34 |
| 225 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | 0.99 | 0.98 | 0.99 | 0.98 | 0.98 | 0.98 | 1.00 | 0.34 |
| 228 | 1.00 | 1.00 | 0.38 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.88 | 1.00 | 1.00 | 0.80 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 230 | 1.00 | 1.00 | 0.71 | 1.00 | 0.94 | 1.00 | 0.91 | 0.97 | 1.00 | 1.00 | 0.88 | 0.96 | 0.94 | 1.00 | 0.97 | 0.85 | 0.94 | 1.00 | 0.86 | 1.00 | 0.94 | 1.00 | 0.94 | 1.00 | 0.94 | 1.00 | 0.88 | 0.96 | 1.00 | 0.35 |
| 231 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 0.34 |
| 232 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 1.00 | 1.00 | 0.65 | 1.00 | 1.00 | 0.94 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 1.00 | 0.34 |
| 233 | 1.00 | 1.00 | 0.38 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.18 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 236 | 1.00 | 1.00 | 0.38 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 0.88 | 1.00 | 1.00 | 0.80 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 |
| 237 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.97 | 0.78 | 1.00 | 1.00 | 0.93 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.97 | 0.31 |
| 238 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 0.95 | 0.95 | 0.99 | 0.99 | 0.97 | 0.97 | 1.00 | 1.00 | 0.85 | 0.78 | 0.99 | 0.99 | 0.90 | 0.95 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.97 | 0.97 | 0.97 | 0.31 |
| 239 | 1.00 | 1.00 | 0.28 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.83 | 0.86 | 0.97 | 1.00 | 0.89 | 0.86 | 1.00 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 |
| 240 | 1.00 | 1.00 | 0.33 | 1.00 | 0.97 | 1.00 | 0.85 | 0.85 | 0.94 | 0.97 | 0.88 | 0.91 | 0.97 | 1.00 | 0.63 | 0.82 | 0.97 | 1.00 | 0.71 | 0.82 | 1.00 | 1.00 | 0.94 | 0.97 | 0.94 | 0.97 | 0.88 | 0.91 | 0.97 | 0.91 |
| 241 | 1.00 | 1.00 | 0.38 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.18 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 246 | 1.00 | 1.00 | 0.28 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.83 | 0.86 | 0.97 | 1.00 | 0.81 | 1.00 | 1.00 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 |
| 247 | 1.00 | 1.00 | 0.33 | 1.00 | 0.97 | 1.00 | 0.85 | 0.85 | 0.94 | 0.97 | 0.88 | 0.91 | 0.97 | 1.00 | 0.63 | 0.82 | 0.94 | 0.97 | 0.73 | 0.82 | 1.00 | 1.00 | 0.94 | 0.97 | 0.94 | 0.97 | 0.88 | 0.91 | 0.97 | 0.91 |
| 248 | 1.00 | 1.00 | 0.03 | 0.03 | 0.50 | 0.12 | 0.92 | 0.36 | 0.91 | 0.73 | n/a | n/a | 1.00 | 0.24 | 0.03 | 0.06 | 1.00 | 0.81 | 0.04 | 0.04 | 1.00 | 0.75 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 248-2 | 1.00 | 1.00 | 0.76 | 0.79 | 0.99 | 0.78 | 0.99 | 0.86 | 0.99 | 0.95 | 1.00 | 0.80 | 0.98 | 0.82 | 0.70 | 0.62 | 1.00 | 0.95 | 0.75 | 0.79 | 1.00 | 0.96 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.80 | 0.88 | 0.22 |
| 248-4 | 1.00 | 1.00 | 0.57 | 0.60 | 0.97 | 0.64 | 0.95 | 0.72 | 0.96 | 0.90 | 1.00 | 0.61 | 0.99 | 0.69 | 0.91 | 0.53 | 1.00 | 0.92 | 0.48 | 0.54 | 1.00 | 0.91 | 1.00 | 0.59 | 1.00 | 0.59 | 1.00 | 0.61 | 0.94 | 0.18 |
| 248-6 | 1.00 | 1.00 | 0.40 | 0.41 | 0.94 | 0.51 | 0.95 | 0.62 | 0.93 | 0.82 | 1.00 | 0.42 | 0.98 | 0.56 | 0.93 | 0.42 | 1.00 | 0.88 | 0.36 | 0.40 | 1.00 | 0.86 | 1.00 | 0.41 | 1.00 | 0.41 | 1.00 | 0.42 | 0.92 | 0.11 |
| 248-8 | 1.00 | 1.00 | 0.23 | 0.24 | 0.74 | 0.32 | 0.88 | 0.45 | 0.93 | 0.80 | 0.95 | 0.21 | 0.95 | 0.40 | 0.49 | 0.32 | 0.98 | 0.85 | 0.16 | 0.18 | 1.00 | 0.84 | 1.00 | 0.22 | 1.00 | 0.22 | 0.95 | 0.21 | 0.83 | 0.05 |
| 249 | 1.00 | 1.00 | 0.03 | 0.03 | 0.92 | 0.24 | 0.17 | 0.01 | 0.84 | 0.71 | n/a | n/a | 1.00 | 0.01 | 0.14 | 0.04 | 0.83 | 0.66 | 0.06 | 0.07 | 0.89 | 0.60 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 249-2 | 1.00 | 1.00 | 0.76 | 0.79 | 1.00 | 0.86 | 0.97 | 0.79 | 0.99 | 0.95 | 1.00 | 0.82 | 0.97 | 0.79 | 0.74 | 0.77 | 0.98 | 0.95 | 0.73 | 0.82 | 1.00 | 0.94 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.82 | 0.84 | 0.22 |
| 249-4 | 1.00 | 1.00 | 0.57 | 0.60 | 1.00 | 0.90 | 0.95 | 0.63 | 0.94 | 0.91 | 1.00 | 0.66 | 0.97 | 0.60 | 0.46 | 0.75 | 0.98 | 0.91 | 0.53 | 0.59 | 1.00 | 0.89 | 1.00 | 0.60 | 1.00 | 0.60 | 1.00 | 0.66 | 0.89 | 0.18 |
| 249-6 | 1.00 | 1.00 | 0.40 | 0.41 | 0.93 | 0.58 | 0.98 | 0.47 | 0.91 | 0.84 | 0.96 | 0.46 | 0.95 | 0.41 | 0.94 | 0.60 | 0.98 | 0.87 | 0.34 | 0.38 | 0.97 | 0.78 | 1.00 | 0.41 | 1.00 | 0.41 | 0.96 | 0.46 | 0.85 | 0.11 |
| 249-8 | 1.00 | 1.00 | 0.22 | 0.23 | 0.95 | 0.77 | 0.90 | 0.28 | 0.87 | 0.75 | 1.00 | 0.27 | 0.88 | 0.22 | 0.77 | 0.48 | 0.95 | 0.82 | 0.16 | 0.18 | 0.91 | 0.69 | 1.00 | 0.22 | 1.00 | 0.22 | 1.00 | 0.27 | 0.83 | 0.05 |
| 250 | 1.00 | 1.00 | 0.01 | 0.03 | 1.00 | 0.33 | 1.00 | 0.36 | 1.00 | 0.36 | n/a | n/a | 1.00 | 0.27 | 0.17 | 0.18 | 0.90 | 0.58 | 0.07 | 0.09 | 1.00 | 0.55 | NaN | 0.00 | NaN | 0.00 | n/a | n/a | NaN | 0.00 |
| 250-2 | 1.00 | 1.00 | 0.30 | 0.79 | 1.00 | 0.85 | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | 0.85 | 0.96 | 0.82 | 0.97 | 0.85 | 1.00 | 1.00 | 0.78 | 0.85 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.85 | 0.84 | 0.64 |
| 250-4 | 1.00 | 1.00 | 0.23 | 0.61 | 1.00 | 0.73 | 1.00 | 0.82 | 1.00 | 1.00 | 1.00 | 0.79 | 1.00 | 0.70 | 0.88 | 0.88 | 1.00 | 1.00 | 0.67 | 0.48 | 1.00 | 1.00 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.79 | 0.89 | 0.52 |
| 250-6 | 1.00 | 1.00 | 0.16 | 0.42 | 1.00 | 0.64 | 1.00 | 0.73 | 1.00 | 0.94 | 0.90 | 0.55 | 1.00 | 0.61 | 0.68 | 0.85 | 1.00 | 1.00 | 0.38 | 0.48 | 1.00 | 0.94 | 1.00 | 0.42 | 1.00 | 0.42 | 0.90 | 0.55 | 0.85 | 0.33 |
| 250-8 | 1.00 | 1.00 | 0.08 | 0.21 | 1.00 | 0.48 | 1.00 | 0.61 | 1.00 | 0.76 | 1.00 | 0.36 | 1.00 | 0.42 | 0.50 | 0.67 | 1.00 | 0.88 | 0.21 | 0.27 | 0.96 | 0.82 | 1.00 | 0.21 | 1.00 | 0.21 | 1.00 | 0.36 | 0.83 | 0.15 |
| 251 | 1.00 | 1.00 | 0.04 | 0.04 | 1.00 | 0.32 | 0.95 | 0.40 | 0.89 | 0.73 | n/a | n/a | 1.00 | 0.25 | 0.17 | 0.06 | 0.96 | 0.76 | 0.07 | 0.08 | 0.76 | 0.59 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 251-2 | 1.00 | 1.00 | 0.73 | 0.80 | 1.00 | 0.85 | 0.98 | 0.86 | 0.98 | 0.95 | 1.00 | 0.83 | 0.99 | 0.84 | 0.97 | 0.70 | 0.99 | 0.96 | 0.76 | 0.80 | 0.98 | 0.94 | 1.00 | 0.80 | 1.00 | 0.80 | 1.00 | 0.83 | 0.82 | 0.19 |
| 251-4 | 1.00 | 1.00 | 0.55 | 0.60 | 1.00 | 0.76 | 0.96 | 0.76 | 0.92 | 0.89 | 1.00 | 0.66 | 0.97 | 0.69 | 0.88 | 0.63 | 0.99 | 0.90 | 0.47 | 0.53 | 0.95 | 0.85 | 1.00 | 0.60 | 1.00 | 0.60 | 1.00 | 0.66 | 0.94 | 0.16 |
| 251-6 | 1.00 | 1.00 | 0.38 | 0.41 | 0.97 | 0.62 | 0.97 | 0.66 | 0.89 | 0.84 | 0.95 | 0.43 | 0.98 | 0.56 | 0.83 | 0.56 | 0.94 | 0.83 | 0.28 | 0.30 | 0.91 | 0.76 | 1.00 | 0.41 | 1.00 | 0.41 | 0.95 | 0.43 | 0.90 | 0.10 |
| 251-8 | 1.00 | 1.00 | 0.22 | 0.24 | 0.92 | 0.47 | 0.92 | 0.52 | 0.90 | 0.81 | 0.96 | 0.25 | 0.95 | 0.42 | 0.47 | 0.46 | 0.99 | 0.85 | 0.22 | 0.24 | 0.87 | 0.71 | 1.00 | 0.23 | 1.00 | 0.23 | 0.96 | 0.25 | 0.83 | 0.05 |
| 252 | 1.00 | 1.00 | 0.02 | 0.02 | 0.80 | 0.08 | 0.88 | 0.38 | 0.91 | 0.75 | n/a | n/a | 0.96 | 0.23 | 0.08 | 0.03 | 0.96 | 0.79 | 0.06 | 0.06 | 0.86 | 0.68 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 252-2 | 1.00 | 1.00 | 0.76 | 0.79 | 0.97 | 0.79 | 0.94 | 0.85 | 0.97 | 0.95 | 0.99 | 0.81 | 0.98 | 0.82 | 0.70 | 0.75 | 0.97 | 0.94 | 0.62 | 0.70 | 0.97 | 0.94 | 0.99 | 0.79 | 0.99 | 0.79 | 0.99 | 0.81 | 0.84 | 0.22 |
| 252-4 | 1.00 | 1.00 | 0.76 | 0.79 | 0.97 | 0.79 | 0.94 | 0.85 | 0.97 | 0.95 | 0.99 | 0.81 | 0.96 | 0.81 | 0.70 | 0.75 | 0.97 | 0.94 | 0.63 | 0.71 | 0.97 | 0.94 | 0.99 | 0.78 | 0.99 | 0.78 | 0.99 | 0.81 | 0.84 | 0.22 |
| 252-6 | 1.00 | 1.00 | 0.76 | 0.79 | 0.98 | 0.80 | 0.94 | 0.85 | 0.97 | 0.95 | 0.98 | 0.81 | 0.98 | 0.82 | 0.70 | 0.75 | 0.97 | 0.94 | 0.63 | 0.69 | 0.97 | 0.94 | 0.99 | 0.79 | 0.99 | 0.79 | 0.98 | 0.81 | 0.84 | 0.22 |
| 252-8 | 1.00 | 1.00 | 0.76 | 0.79 | 0.98 | 0.80 | 0.94 | 0.85 | 0.97 | 0.95 | 0.99 | 0.81 | 0.98 | 0.82 | 0.70 | 0.75 | 0.97 | 0.94 | 0.63 | 0.71 | 0.97 | 0.94 | 0.99 | 0.79 | 0.99 | 0.79 | 0.99 | 0.81 | 0.84 | 0.22 |
| 253 | 1.00 | 1.00 | 0.03 | 0.03 | 0.46 | 0.11 | 0.17 | 0.01 | 0.83 | 0.59 | n/a | n/a | 1.00 | 0.01 | 0.03 | 0.04 | 0.81 | 0.59 | 0.06 | 0.07 | 0.87 | 0.46 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 253-2 | 1.00 | 1.00 | 0.76 | 0.79 | 0.99 | 0.78 | 0.97 | 0.77 | 0.99 | 0.93 | 1.00 | 0.80 | 0.97 | 0.79 | 0.70 | 0.62 | 0.98 | 0.93 | 0.75 | 0.71 | 1.00 | 0.92 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.80 | 0.88 | 0.22 |
| 253-4 | 1.00 | 1.00 | 0.57 | 0.60 | 0.99 | 0.68 | 0.92 | 0.57 | 0.93 | 0.87 | 1.00 | 0.61 | 0.97 | 0.60 | 0.91 | 0.53 | 1.00 | 0.92 | 0.50 | 0.56 | 1.00 | 0.87 | 1.00 | 0.60 | 1.00 | 0.60 | 1.00 | 0.61 | 0.94 | 0.18 |
| 253-6 | 1.00 | 1.00 | 0.40 | 0.41 | 0.93 | 0.52 | 0.93 | 0.41 | 0.93 | 0.78 | 1.00 | 0.42 | 0.95 | 0.41 | 0.93 | 0.42 | 0.95 | 0.81 | 0.38 | 0.42 | 0.97 | 0.77 | 1.00 | 0.41 | 1.00 | 0.41 | 1.00 | 0.42 | 0.92 | 0.11 |
| 253-8 | 1.00 | 1.00 | 0.23 | 0.24 | 0.79 | 0.34 | 0.76 | 0.20 | 0.89 | 0.72 | 0.95 | 0.21 | 0.88 | 0.22 | 0.49 | 0.32 | 0.95 | 0.79 | 0.17 | 0.19 | 0.90 | 0.66 | 1.00 | 0.22 | 1.00 | 0.22 | 0.95 | 0.21 | 0.83 | 0.05 |
| 254 | 1.00 | 1.00 | 0.01 | 0.03 | 1.00 | 0.27 | 0.89 | 0.24 | 1.00 | 0.27 | n/a | n/a | 1.00 | 0.27 | 0.03 | 0.18 | 1.00 | 0.27 | 0.00 | 0.00 | 1.00 | 0.27 | NaN | 0.00 | NaN | 0.00 | n/a | n/a | NaN | 0.00 |
| 254-2 | 1.00 | 1.00 | 0.30 | 0.79 | 1.00 | 0.82 | 1.00 | 0.82 | 1.00 | 0.82 | 1.00 | 0.79 | 0.96 | 0.82 | 0.10 | 0.18 | 1.00 | 0.82 | 0.85 | 0.70 | 1.00 | 0.82 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.79 | 0.88 | 0.64 |
| 254-4 | 1.00 | 1.00 | 0.23 | 0.61 | 1.00 | 0.70 | 0.91 | 0.64 | 1.00 | 0.70 | 1.00 | 0.61 | 1.00 | 0.70 | 0.06 | 0.18 | 1.00 | 0.70 | 0.83 | 0.45 | 1.00 | 0.70 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.61 | 0.94 | 0.52 |
| 254-6 | 1.00 | 1.00 | 0.16 | 0.42 | 1.00 | 0.61 | 0.90 | 0.55 | 1.00 | 0.61 | 1.00 | 0.42 | 1.00 | 0.61 | 0.06 | 0.18 | 1.00 | 0.61 | 0.37 | 0.39 | 1.00 | 0.61 | 1.00 | 0.42 | 1.00 | 0.42 | 1.00 | 0.42 | 0.92 | 0.33 |
| 254-8 | 1.00 | 1.00 | 0.08 | 0.21 | 1.00 | 0.42 | 0.71 | 0.30 | 1.00 | 0.42 | 1.00 | 0.21 | 1.00 | 0.42 | 0.05 | 0.18 | 1.00 | 0.42 | 0.71 | 0.15 | 1.00 | 0.42 | 1.00 | 0.21 | 1.00 | 0.21 | 1.00 | 0.21 | 0.83 | 0.15 |
| 257 | 1.00 | 1.00 | 0.01 | 0.03 | 0.50 | 0.06 | NaN | 0.00 | 0.50 | 0.06 | n/a | n/a | NaN | 0.00 | 0.17 | 0.18 | 0.50 | 0.06 | 0.05 | 0.06 | 1.00 | 0.18 | NaN | 0.00 | NaN | 0.00 | n/a | n/a | NaN | 0.00 |
| 257-2 | 1.00 | 1.00 | 0.30 | 0.79 | 1.00 | 0.82 | 1.00 | 0.85 | 1.00 | 0.97 | 1.00 | 0.85 | 0.96 | 0.79 | 0.97 | 0.85 | 1.00 | 0.97 | 0.91 | 0.61 | 1.00 | 0.97 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.85 | 0.84 | 0.64 |
| 257-4 | 1.00 | 1.00 | 0.23 | 0.61 | 0.95 | 0.64 | 1.00 | 0.73 | 1.00 | 0.88 | 1.00 | 0.76 | 1.00 | 0.61 | 0.88 | 0.88 | 0.94 | 0.88 | 0.53 | 0.61 | 1.00 | 0.88 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.76 | 0.89 | 0.52 |
| 257-6 | 1.00 | 1.00 | 0.16 | 0.42 | 0.88 | 0.45 | 1.00 | 0.55 | 0.92 | 0.67 | 0.90 | 0.55 | 0.93 | 0.42 | 0.68 | 0.85 | 0.84 | 0.79 | 0.40 | 0.52 | 1.00 | 0.70 | 1.00 | 0.42 | 1.00 | 0.42 | 0.90 | 0.55 | 0.85 | 0.33 |
| 257-8 | 1.00 | 1.00 | 0.09 | 0.24 | 0.82 | 0.27 | 1.00 | 0.36 | 0.88 | 0.45 | 1.00 | 0.36 | 0.88 | 0.21 | 0.50 | 0.67 | 0.89 | 0.76 | 0.23 | 0.27 | 1.00 | 0.52 | 1.00 | 0.21 | 1.00 | 0.21 | 1.00 | 0.36 | 0.83 | 0.15 |
| 258 | 1.00 | 1.00 | 0.04 | 0.04 | 1.00 | 0.31 | 0.17 | 0.01 | 0.71 | 0.55 | n/a | n/a | 1.00 | 0.01 | 0.13 | 0.03 | 0.80 | 0.60 | 0.08 | 0.09 | 0.40 | 0.22 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 258-2 | 1.00 | 1.00 | 0.73 | 0.80 | 1.00 | 0.86 | 0.96 | 0.77 | 0.98 | 0.92 | 0.99 | 0.82 | 0.97 | 0.80 | 0.97 | 0.70 | 0.97 | 0.94 | 0.74 | 0.74 | 0.98 | 0.89 | 1.00 | 0.78 | 1.00 | 0.78 | 0.99 | 0.82 | 0.82 | 0.19 |
| 258-4 | 1.00 | 1.00 | 0.55 | 0.60 | 1.00 | 0.76 | 0.93 | 0.60 | 0.92 | 0.87 | 1.00 | 0.66 | 0.97 | 0.60 | 0.88 | 0.63 | 0.96 | 0.88 | 0.49 | 0.53 | 0.93 | 0.76 | 1.00 | 0.59 | 1.00 | 0.59 | 1.00 | 0.66 | 0.94 | 0.16 |
| 258-6 | 1.00 | 1.00 | 0.38 | 0.41 | 0.97 | 0.62 | 0.95 | 0.44 | 0.87 | 0.78 | 0.93 | 0.43 | 0.95 | 0.41 | 0.81 | 0.56 | 0.95 | 0.82 | 0.34 | 0.39 | 0.85 | 0.62 | 1.00 | 0.41 | 1.00 | 0.41 | 0.93 | 0.43 | 0.90 | 0.10 |
| 258-8 | 1.00 | 1.00 | 0.22 | 0.24 | 0.91 | 0.46 | 0.81 | 0.24 | 0.76 | 0.65 | 1.00 | 0.26 | 0.88 | 0.23 | 0.47 | 0.46 | 0.94 | 0.78 | 0.20 | 0.23 | 0.77 | 0.51 | 1.00 | 0.23 | 1.00 | 0.23 | 1.00 | 0.26 | 0.83 | 0.05 |
| 259 | 1.00 | 1.00 | 0.03 | 0.03 | 0.11 | 0.01 | 0.50 | 0.06 | 0.75 | 0.61 | n/a | n/a | 0.88 | 0.07 | 0.09 | 0.03 | 0.89 | 0.70 | 0.01 | 0.01 | 0.70 | 0.41 | 1.00 | 0.01 | 1.00 | 0.01 | n/a | n/a | NaN | 0.00 |
| 259-2 | 1.00 | 1.00 | 0.76 | 0.79 | 0.98 | 0.80 | 0.91 | 0.77 | 0.97 | 0.93 | 0.98 | 0.81 | 0.96 | 0.80 | 0.70 | 0.75 | 0.98 | 0.95 | 0.68 | 0.76 | 0.96 | 0.91 | 0.99 | 0.78 | 0.99 | 0.78 | 0.98 | 0.81 | 0.84 | 0.22 |
| 259-4 | 1.00 | 1.00 | 0.76 | 0.79 | 0.98 | 0.80 | 0.91 | 0.77 | 0.97 | 0.93 | 0.98 | 0.81 | 0.96 | 0.80 | 0.70 | 0.75 | 0.98 | 0.95 | 0.64 | 0.72 | 0.96 | 0.91 | 0.99 | 0.79 | 0.99 | 0.79 | 0.98 | 0.81 | 0.84 | 0.22 |
| 259-6 | 1.00 | 1.00 | 0.76 | 0.79 | 0.98 | 0.80 | 0.91 | 0.77 | 0.97 | 0.93 | 0.99 | 0.81 | 0.96 | 0.80 | 0.70 | 0.75 | 0.98 | 0.95 | 0.66 | 0.74 | 0.96 | 0.91 | 0.99 | 0.78 | 0.99 | 0.78 | 0.99 | 0.81 | 0.84 | 0.22 |
| 259-8 | 1.00 | 1.00 | 0.76 | 0.79 | 0.97 | 0.79 | 0.91 | 0.77 | 0.97 | 0.93 | 0.99 | 0.81 | 0.96 | 0.80 | 0.70 | 0.75 | 0.98 | 0.95 | 0.66 | 0.73 | 0.96 | 0.91 | 0.99 | 0.79 | 0.99 | 0.79 | 0.99 | 0.81 | 0.84 | 0.22 |
| 260 | 1.00 | 1.00 | 0.02 | 0.07 | 0.92 | 0.38 | 1.00 | 0.34 | 0.92 | 0.41 | n/a | n/a | 0.90 | 0.31 | 0.17 | 0.21 | 0.94 | 0.55 | 0.03 | 0.03 | 0.93 | 0.45 | 0.00 | 0.00 | 0.00 | 0.00 | n/a | n/a | NaN | 0.00 |
| 260-2 | 1.00 | 1.00 | 0.23 | 0.79 | 0.96 | 0.86 | 0.96 | 0.83 | 0.96 | 0.90 | 0.96 | 0.83 | 0.92 | 0.83 | 0.83 | 0.83 | 0.96 | 0.93 | 0.67 | 0.76 | 1.00 | 0.93 | 0.96 | 0.79 | 0.96 | 0.79 | 0.96 | 0.83 | 0.82 | 0.62 |
| 260-4 | 1.00 | 1.00 | 0.18 | 0.62 | 0.96 | 0.76 | 0.96 | 0.76 | 0.96 | 0.86 | 0.91 | 0.72 | 0.95 | 0.72 | 0.63 | 0.83 | 0.93 | 0.93 | 0.53 | 0.72 | 1.00 | 0.90 | 0.95 | 0.62 | 0.95 | 0.62 | 0.91 | 0.72 | 0.94 | 0.52 |
| 260-6 | 1.00 | 1.00 | 0.12 | 0.41 | 0.95 | 0.66 | 0.95 | 0.66 | 0.96 | 0.76 | 0.78 | 0.48 | 0.95 | 0.62 | 0.45 | 0.62 | 0.96 | 0.79 | 0.64 | 0.31 | 1.00 | 0.79 | 0.92 | 0.41 | 0.92 | 0.41 | 0.78 | 0.48 | 0.90 | 0.31 |
| 260-8 | 1.00 | 1.00 | 0.08 | 0.28 | 0.94 | 0.55 | 0.88 | 0.48 | 0.95 | 0.66 | 0.91 | 0.34 | 0.93 | 0.48 | 0.16 | 0.72 | 0.88 | 0.72 | 0.21 | 0.28 | 1.00 | 0.72 | 0.88 | 0.24 | 0.88 | 0.24 | 0.91 | 0.34 | 0.83 | 0.17 |
| 261 | 1.00 | 1.00 | 0.01 | 0.03 | 0.82 | 0.27 | 0.77 | 0.30 | 0.85 | 0.33 | n/a | n/a | 0.80 | 0.24 | 0.08 | 0.09 | 0.67 | 0.48 | 0.04 | 0.06 | 1.00 | 0.27 | 0.00 | 0.00 | 0.00 | 0.00 | n/a | n/a | NaN | 0.00 |
| 261-2 | 1.00 | 1.00 | 0.26 | 0.79 | 0.96 | 0.82 | 0.90 | 0.82 | 0.88 | 0.91 | 0.94 | 0.88 | 0.93 | 0.82 | 0.70 | 0.79 | 0.88 | 0.91 | 0.86 | 0.36 | 0.90 | 0.79 | 0.93 | 0.79 | 0.93 | 0.79 | 0.94 | 0.88 | 0.84 | 0.64 |
| 261-4 | 1.00 | 1.00 | 0.26 | 0.79 | 0.97 | 0.85 | 0.90 | 0.82 | 0.88 | 0.91 | 0.93 | 0.85 | 0.93 | 0.82 | 0.70 | 0.79 | 0.88 | 0.91 | 0.82 | 0.27 | 0.90 | 0.79 | 0.93 | 0.79 | 0.93 | 0.79 | 0.93 | 0.85 | 0.84 | 0.64 |
| 261-6 | 1.00 | 1.00 | 0.26 | 0.79 | 0.97 | 0.85 | 0.90 | 0.82 | 0.88 | 0.91 | 0.93 | 0.85 | 0.90 | 0.79 | 0.70 | 0.79 | 0.88 | 0.91 | 0.75 | 0.45 | 0.90 | 0.79 | 0.93 | 0.79 | 0.93 | 0.79 | 0.93 | 0.85 | 0.84 | 0.64 |
| 261-8 | 1.00 | 1.00 | 0.26 | 0.79 | 0.96 | 0.82 | 0.90 | 0.82 | 0.88 | 0.91 | 0.90 | 0.85 | 0.90 | 0.79 | 0.70 | 0.79 | 0.88 | 0.91 | 0.68 | 0.79 | 0.90 | 0.79 | 0.93 | 0.79 | 0.93 | 0.79 | 0.90 | 0.85 | 0.84 | 0.64 |
| 262 | 1.00 | 1.00 | 0.01 | 0.03 | NaN | 0.00 | NaN | 0.00 | NaN | 0.00 | n/a | n/a | NaN | 0.00 | 0.03 | 0.18 | NaN | 0.00 | 0.07 | 0.09 | NaN | 0.00 | NaN | 0.00 | NaN | 0.00 | n/a | n/a | NaN | 0.00 |
| 262-2 | 1.00 | 1.00 | 0.30 | 0.79 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.76 | 1.00 | 0.79 | 0.96 | 0.79 | 0.10 | 0.18 | 1.00 | 0.79 | 0.86 | 0.76 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.79 | 1.00 | 0.79 | 0.88 | 0.64 |
| 262-4 | 1.00 | 1.00 | 0.23 | 0.61 | 1.00 | 0.61 | 0.90 | 0.55 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.61 | 0.06 | 0.18 | 1.00 | 0.61 | 0.50 | 0.55 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.61 | 1.00 | 0.61 | 0.94 | 0.52 |
| 262-6 | 1.00 | 1.00 | 0.16 | 0.42 | 1.00 | 0.42 | 0.86 | 0.36 | 1.00 | 0.42 | 1.00 | 0.42 | 0.93 | 0.42 | 0.06 | 0.18 | 1.00 | 0.42 | 0.79 | 0.33 | 1.00 | 0.42 | 1.00 | 0.42 | 1.00 | 0.42 | 1.00 | 0.42 | 0.92 | 0.33 |
| 262-8 | 1.00 | 1.00 | 0.08 | 0.21 | 1.00 | 0.21 | 0.57 | 0.12 | 1.00 | 0.21 | 1.00 | 0.21 | 0.88 | 0.21 | 0.05 | 0.18 | 1.00 | 0.21 | 0.16 | 0.21 | 1.00 | 0.21 | 1.00 | 0.21 | 1.00 | 0.21 | 1.00 | 0.21 | 0.83 | 0.15 |
| 265 | 1.00 | 1.00 | 0.02 | 0.07 | 0.50 | 0.07 | NaN | 0.00 | 0.40 | 0.07 | n/a | n/a | 0.00 | 0.00 | 0.17 | 0.21 | 0.80 | 0.14 | 0.03 | 0.03 | 0.75 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | n/a | n/a | NaN | 0.00 |
| 266 | 1.00 | 1.00 | 0.01 | 0.03 | 0.25 | 0.03 | 0.00 | 0.00 | 0.40 | 0.06 | n/a | n/a | 0.00 | 0.00 | 0.08 | 0.09 | 0.30 | 0.09 | 0.02 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | n/a | n/a | NaN | 0.00 |
| 301 | 1.00 | 1.00 | 0.48 | 0.79 | 0.98 | 0.82 | 0.84 | 0.67 | 0.89 | 0.77 | 0.88 | 0.59 | 0.87 | 0.43 | 0.67 | 0.54 | 0.94 | 0.82 | NaN | 0.00 | 0.76 | 0.69 | 0.96 | 0.79 | 0.96 | 0.79 | 0.27 | 0.67 | 1.00 | 0.21 |
| 302 | 1.00 | 1.00 | 0.31 | 0.65 | 0.89 | 0.65 | 0.79 | 0.54 | 0.61 | 0.46 | 0.94 | 0.60 | 0.90 | 0.58 | 0.40 | 0.44 | 0.89 | 0.65 | 0.22 | 0.21 | 0.72 | 0.65 | 1.00 | 0.60 | 0.97 | 0.60 | 0.26 | 0.75 | 0.91 | 0.21 |
| 303 | 1.00 | 1.00 | 0.40 | 0.83 | n/a | n/a | 0.73 | 0.60 | 0.73 | 0.83 | 0.81 | 0.79 | 0.80 | 0.81 | NaN | 0.00 | 0.65 | 0.71 | NaN | 0.00 | 0.76 | 0.88 | 0.89 | 0.83 | 0.78 | 0.83 | 0.08 | 0.81 | n/a | n/a |
| 304 | 1.00 | 1.00 | 0.71 | 0.95 | 0.96 | 0.96 | 0.88 | 0.92 | 0.90 | 0.92 | 0.95 | 0.95 | 0.97 | 0.95 | 0.76 | 0.51 | 0.95 | 0.97 | 0.65 | 0.64 | 0.90 | 0.97 | 0.96 | 0.92 | 0.95 | 0.93 | 0.16 | 0.95 | 0.89 | 0.33 |
| H-mean | 1.00 | 1.00 | 0.43 | 0.59 | 0.97 | 0.71 | 0.95 | 0.70 | 0.95 | 0.86 | 0.97 | 0.62 | 0.97 | 0.67 | 0.60 | 0.58 | 0.97 | 0.88 | 0.51 | 0.54 | 0.96 | 0.84 | 0.99 | 0.58 | 0.98 | 0.59 | 0.81 | 0.63 | 0.91 | 0.22 |
n/a: result alignment not provided or not readable
NaN: division per zero, likely due to empty alignent.