We have run the evaluation in a Ubuntu Laptop with an Intel Core i7-4600U CPU @ 2.10GHz x 4 and allocating 15Gb of RAM.
Systems have been evaluated according to the following criteria:
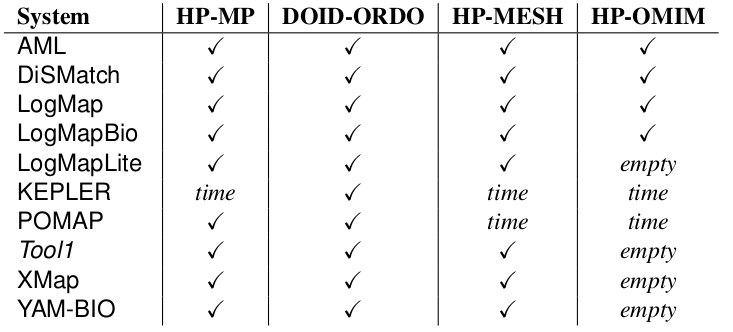
In the OAEI 2017 phenotype track 10 out of 21 participating OAEI 2017 systems have been able to cope with at least one of the tasks with a 4 hours timeout (see Table 1).
LogMapBio uses BioPortal as mediating ontology provider, that is, it retrieves from BioPortal the most suitable top-10 ontologies for the matching task.
LogMap uses normalisations and spelling variants from the general (biomedical) purpose UMLS Lexicon.
AML has three sources of background knowledge which can be used as mediators between the input ontologies: the Uber Anatomy Ontology (Uberon), the Human Disease Ontology (DOID) and the Medical Subject Headings (MeSH).
YAM-BIO uses as background knowledge a file containing mappings from the DOID and UBERON ontologies to other ontologies like FMA, NCI or SNOMED CT.
XMAP uses synonyms provided by the UMLS Metathesaurus.
1. Results against the consensus alignments with vote 2, 3 and 4
Table 2 shows the size of the consensus alignments built with the outputs of the systems participating in the OAEI 2016 and 2017 campaigns. Note that systems participating with different variants only contributed once in the voting, that is, the voting was done by family of systems/variants rather than by individual systems.| Task | Vote 2 | Vote 3 | Vote 4 |
|---|---|---|---|
| HP-MP | 3,130 | 2,153 | 1,780 |
| DOID-ORDO | 3,354 | 2,645 | 2,188 |
| HP-MESH | 4,711 | 3,847 | 3,227 |
| HP-OMIM | 6,834 | 4,177 | 3,462 |
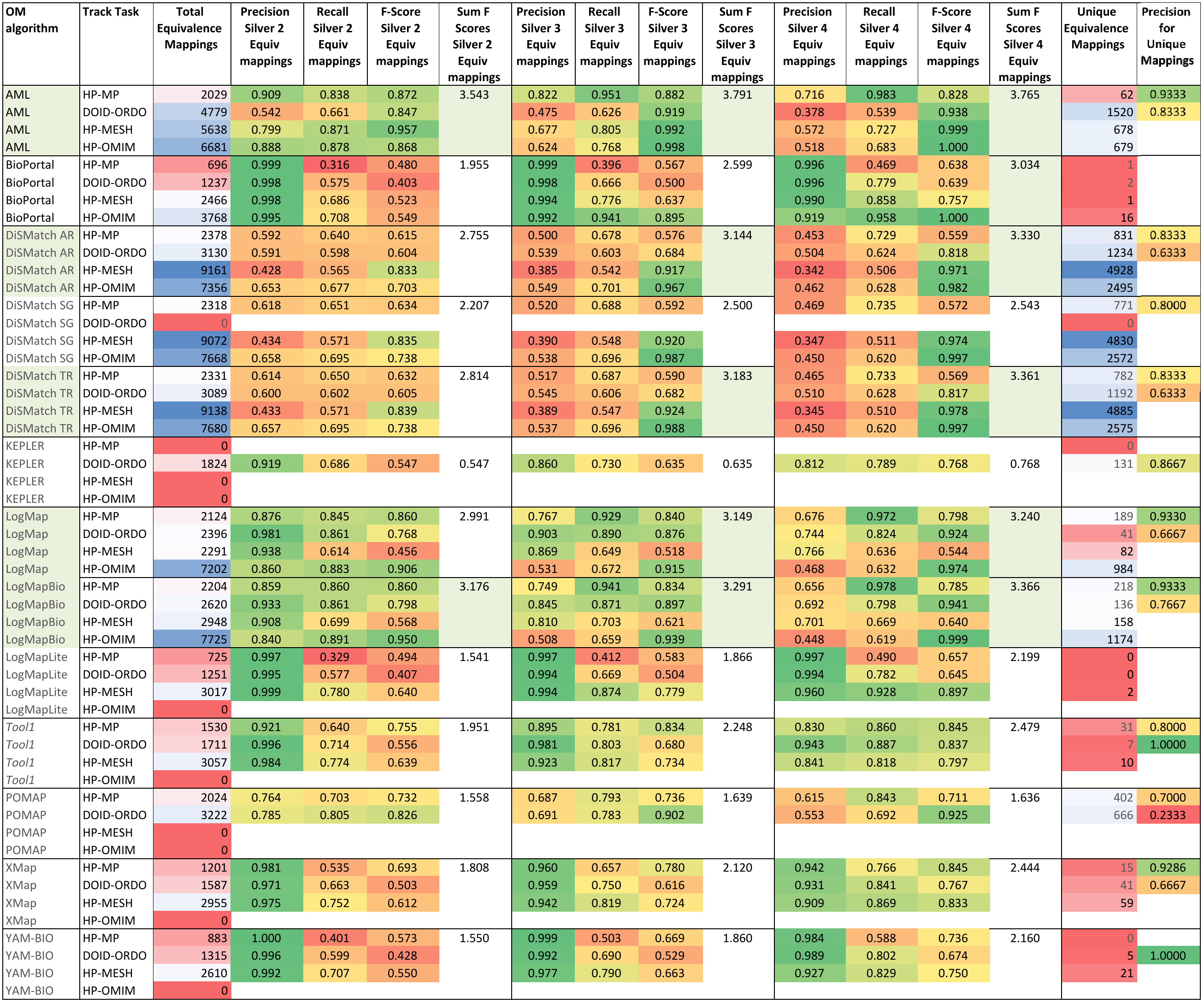
Table 3 shows the results achieved by each of the participating systems. We deliberately did not rank the systems since the consensus alignments only allow us to assess how systems perform in comparison with one another. On the one hand, some of the mappings in the consensus alignment may be erroneous (false positives), as all it takes for that is that 2, 3 or 4 systems agree on part of the erroneous mappings they find. On the other hand, the consensus alignments are not complete, as there will likely be correct mappings that no system is able to find, and as we will show in the manual evaluation, there are a number of mappings found by only one system (and therefore not in the consensus alignments) which are correct. Nevertheless, the results with respect to the consensus alignments do provide some insights into the performance of the systems, which is why we highlighted in the table the 4 systems that produce results closest to the consensus alignments: AML, DiSMatch, LogMap LogMapBio.
2. Results against manually created mappings
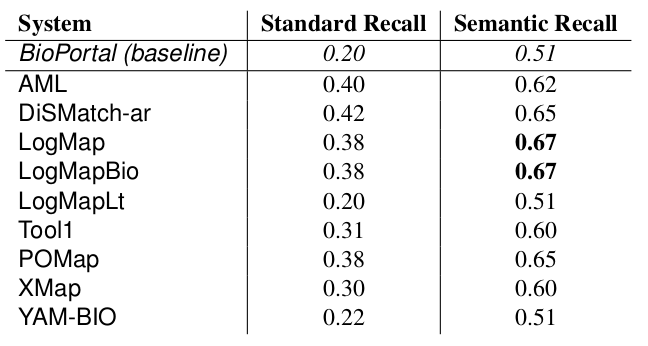
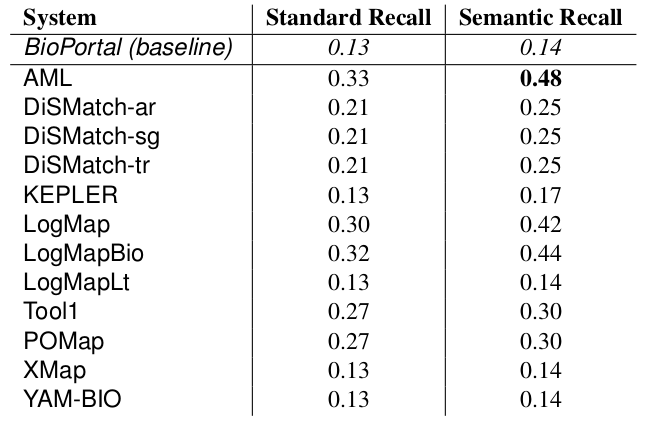
The manually generated mappings for six areas (carbohydrate, obesity and breast cancer, urinary incontinence, abnormal heart and Charcot-Marie Tooth disease) include 86 mappings between HP and MP and 175 mappings between DOID and ORDO. Most of them represent subsumption relationships. Tables 4 and 5 shows the results in terms of recall and semantic recall for each of the system. LogMapBio and LogMap obtained the best results in terms of semantic recall in the HP-MP task, while AML obtained the best results in the DOID-ORDO task. The results in both tasks are far from optimal since a large fragment of the manually created mappings have not been (explicitly) identified by the systems or can be derived via reasoning.
3. Manual assessment of unique mappings
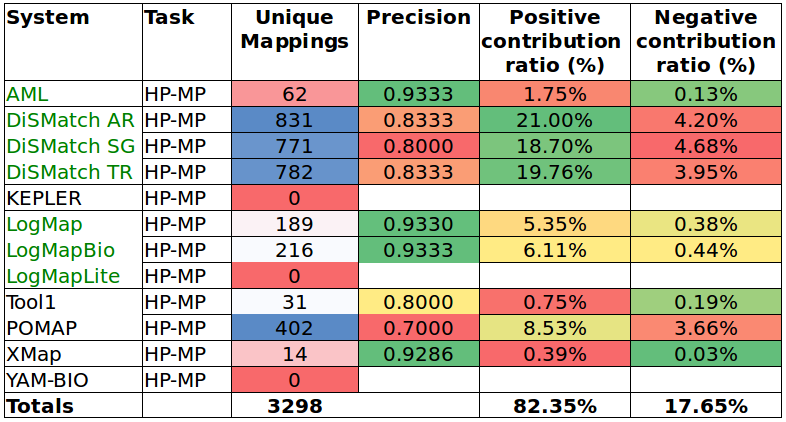
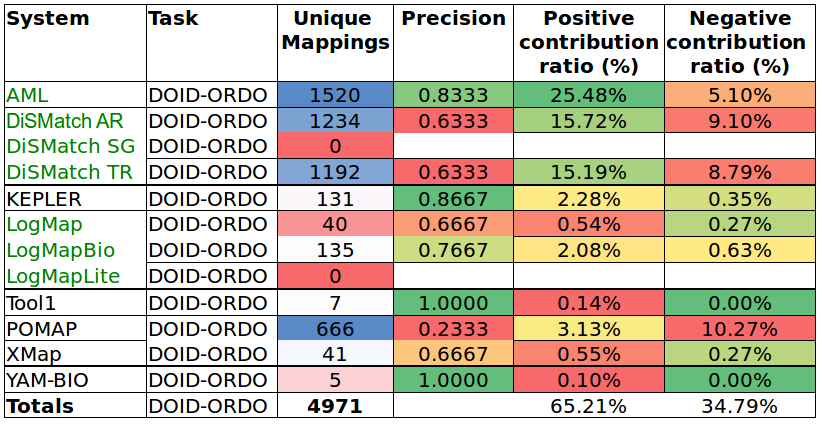
Tables 6 and 7 show the results of the manual assessment to estimate the precision of the unique mappings generated by the participating systems. Unique mappings are correspondences that no other system (explicitly) provided in the output. We manually evaluated up to 30 mappings and we focused the assessment on unique equivalence mappings.
For example LogMap's output contains 189 unique mappings in the HP-MP task. The manual assessment revealed an (estimated) precision of 0.9333. In order to also take into account the number of unique mappings that a system is able to discover, Tables 6 and 7 also include the estimation of the positive and negative contribution of the unique mappings with respect to the total unique mappings discovered by all participating systems.