

In the following are the current results of the OAEI 2016 evaluation of the benchmarks track.
So far, this year we used the test with the usual biblio ontology and a blind newly generated test on a movie ontology provided by IRIT.
In order to avoid the discrepancy of last year, all systems were run in the most simple homogeneous setting. So, this year, we can again write: All tests have been run entirely in the same conditions with the same strict protocol.
Evaluations were run on a Debian Linux virtual machine configured with four processors and 8GB of RAM running under a Dell PowerEdge T610 with 2*Intel Xeon Quad Core 2.26GHz E5607 processors and 32GB of RAM, under Linux ProxMox 2 (Debian). All matchers where run under the SEALS client using Java 1.8 and a maximum heap size of 8GB.
As a result, many systems were not able to properly match the benchmark. Evaluators availability is not unbounded and after running OAEI for more than 10 years, wandering into README files and install software to run evaluations, is not practical. As, reported last year, a good tool is a tool easy to install, i.e., in which the user does not have many reasons to not using it.
We report below the results obtained by all systems:
| Restriction | Version | Errors | Results | F-measure | ||
| Alin | Interactive only | supp _v3 | WordNet | No res. | ||
| AML | .38 | |||||
| CroLOM | WordNet (TO/film) | Empty | ||||
| CroMatch | supp er | .89 | ||||
| DKP-AOM | ArrayIndexOutOfBoundsException | No res. | ||||
| DKP-AOM-Lite | ArrayIndexOutOfBoundsException | No res. | ||||
| DiSMatch | MatcherBridge not found | No res. | ||||
| FCA-Map | IM only | mod _ to - | NullPointerException | No res. | ||
| GA4OM | cannot access configuration file | No res. | ||||
| IOMap | mod IOMAP_v2 | (TO/film) | Empty | |||
| LPHOM | No CPLEX | No res. | ||||
| LYAM | mod Lyam++ LYAM_v2 | StringIndexOutOfBoundsException (TO/film) | No res. | |||
| Lily | .89 | |||||
| LogMap | .55 | |||||
| LogMapIM | IM only | |||||
| LogMapLt | from LogMapLite | .46 | ||||
| NAISC | supp _v2 | Could not find or load main class | No res. | |||
| PhenoMF | .01 | |||||
| PhenoMM | .01 | |||||
| PhenoMP | .01 | |||||
| XMap | .56 | |||||
| LogMapBio | Largebio only | Very long | .32 | |||
| RiMOM | IM only | mod RIMOM_v2 | Does not hang off (TO/film) | Empty |
However we encountered problems with one very slow matcher (LogMapBio) that has been run anyway. RiMOM did not terminate, but was able to provide (empty) alignments for biblio, not for film. No timeout was explicitly set.
Reported figures are the average of 5 runs. As has already been shown in [1], there is not much variance in compliance measures across runs.
From the 23 systems tested, we were so far able to evaluate 10 systems.
| biblio | film | |||||
| Matching system | Prec. | F-m. | Rec. | Prec. | F-m. | Rec. |
| edna | .35(.58) | .41(.54) | .51(.50) | .43 (.68) | .47 (.58) | .50 (.50) |
| AML | 1.0 | .38 | .24 | 1.0 | .32 | .20 |
| CroMatch | .96 (.60) | .89 (.54) | .83 (.50) | NaN | ||
| Lily | .97 (.45) | .89 (.40) | .83 (.36) | .97 (.39) | .81 (.31) | .70 (.26) |
| LogMap | .93 (.90) | .55 (.53) | .39 (.37) | .83 (.79) | .13 (.12) | .07 (.06) |
| LogMapLt | .43 | .46 | .50 | .62 | .51 | .44 |
| PhenoMF | .03 | .01 | .01 | .03 | .01 | .01 |
| PhenoMM | .03 | .01 | .01 | .03 | .01 | .01 |
| PhenoMP | .02 | .01 | .01 | .03 | .01 | .01 |
| XMap | .95 (.98) | .56 (.57) | .40 (.40) | .78 (.84) | .60 (.62) | .49 (.49) |
| LogMapBio | .48 (.48) | .32 (.30) | .24 (.22) | .59 (.58) | .07 (.06) | .03 (.03) |
It is clear that systems that participated previously (AML, CroMatcher, Lily, LogMap, LogMapLite, XMap) still obtain the best results with Lily and CroMatcher still achieving impresive .89 F-measure (against .90 and .88 last year). They combine very high precision (.96 and .97) with high recall (.83). The PhenoXX suite of systems return huge but poor alignments. It is surprising that some of the systems (AML, LogMapLite) do not clearly outperform edna (our edit distance baseline).
On the film data set (which was not known from the participants when submitting their systems, and actually have been generated afterwards), the results of biblio are fully confirmed: (1) those system able to return results were still able to do it besides CroMatch and those unable, were still not able; (2) the order between these systems and their performances are commensurate. Point (1) shows that these are robust systems. Point (2) shows that the performances of these system are consistent across data sets, hence we are indeed measuring something. However, (2) has for exception LogMap and LogMapBio whose precision is roughly preserved but whose recall dramatically drops. A tentative explanation is that film contains many labels in French and these two systems rely too much on WordNet. Anyway, these and CroMatch seem to show some overfit to biblio.
Besides LogMapLite, all systems have higher precision than recall as usual and usually very high precision as shown on the triangle graph for biblio. This can be compared with last year.
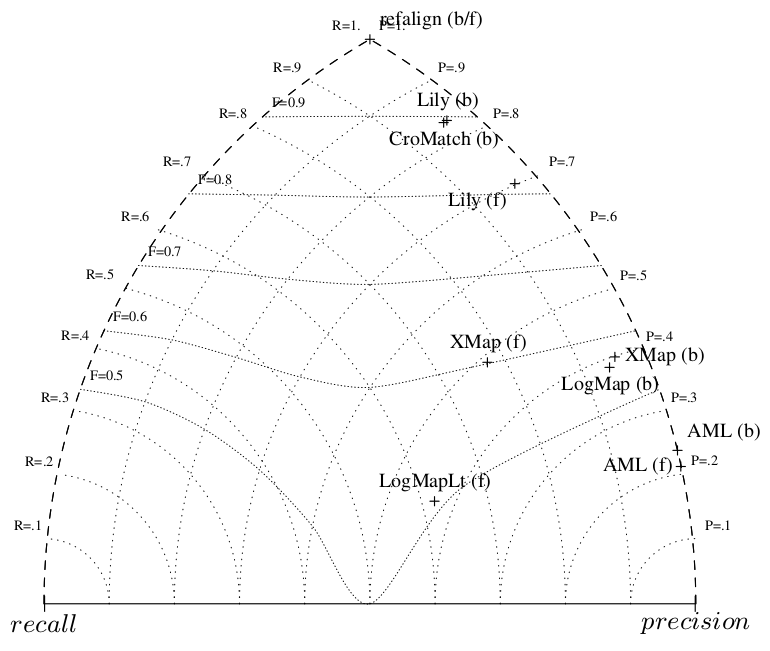
The precision/recall graph confirms that, as usual, there are a level of recall unreachable by any system and this is where some of them go to catch their good F-measure.
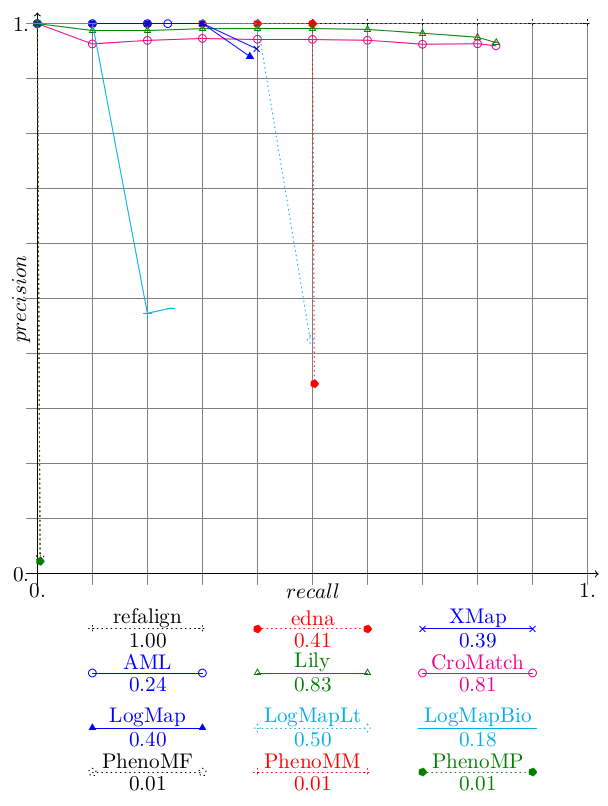
Concerning confidence-weighted measures, there are two types of systems: those (CroMatch, Lily) which obviously threshold their results but keep low confidence values and those (LogMap, XMap, LogMapBio) which provide relatively faithful measures. The former see a strong degradation of the measured values while the latters resist very well with XMap improving its score. This measure which is supposed to reward systems able to provide accurate confidence values is beneficial to these faithful systems.
Beside LogMapBio which uses alignment repositories on the web to find matches, all matchers do the task in less than 40mn (for biblio and 12h for film). There is still a large discrepancy between matchers concerning the time spent from less than two minutes for LogMapLite, AML and XMap to nearly two hours for LogMapBio (on biblio).
| biblio | film | |||||
| Matching system | time (s) | stdev | F-m./s | s. | stdev | F-m./s. |
| AML | 120 | 16 | .32 | 183 | 1 | .17 |
| CroMatch | 1100 | 34 | .08 | NaN | ||
| Lily | 2211 | 31 | .04 | 2797 | 26 | .03 |
| LogMap | 194 | 10 | .28 | 40609 | 13262 | .00 |
| LogMapLt | 96 | 10 | .48 | 116 | 0. | .44 |
| PhenoMF | 1632 | 126 | .00 | 1798 | 123 | .00 |
| PhenoMM | 1743 | 119 | .00 | 1909 | 139 | .00 |
| PhenoMP | 1833 | 123 | .00 | 1835 | 132 | .00 |
| XMap | 123 | 11 | .46 | 2981 | 617 | .02 |
| LogMapBio | 54439 | 3264 | .00 | 193763419 | 61670837 | .00 |
We provide the average time, time standard deviation and 1/100e F-measure point provided per second by matchers. The F-measure point provided per second shows that efficient matchers are, like two years ago, LogMapLite and XMap followed by AML and LogMap. The correlation between time and F-measure only holds for these systems.
You can obtain the detailed results, test by test, by clicking on the test run number.
| algo | refalign | edna | Alin | AML | CroLOM | CroMatch | DKP-AOM | DKP-AOM-Lite | DiSMatch | FCA-Map | GA4OM | IOMap | LPHOM | LYAM | Lily | LogMap | LogMapLt | NAISC | PhenoMF | PhenoMM | PhenoMP | XMap | LogMapBio | RiMOM | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. |
| biblio | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.83 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.89 | 0.83 | 0.93 | 0.55 | 0.39 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.95 | 0.56 | 0.40 | 0.48 | 0.32 | 0.24 | 0.00 | NaN | 0.00 |
| 1 | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.39 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.84 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.90 | 0.84 | 0.93 | 0.57 | 0.41 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.95 | 0.56 | 0.40 | 0.48 | 0.31 | 0.23 | 0.00 | NaN | 0.00 |
| 2 | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.39 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.83 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.83 | 0.93 | 0.55 | 0.39 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.95 | 0.56 | 0.40 | 0.47 | 0.31 | 0.23 | 0.00 | NaN | 0.00 |
| 3 | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.39 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.84 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.90 | 0.84 | 0.93 | 0.56 | 0.40 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.04 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.95 | 0.56 | 0.39 | 0.49 | 0.32 | 0.24 | 0.00 | NaN | 0.00 |
| 4 | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.83 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.89 | 0.83 | 0.93 | 0.54 | 0.38 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.95 | 0.56 | 0.40 | 0.48 | 0.33 | 0.25 | 0.00 | NaN | 0.00 |
| 5 | 1.00 | 1.00 | 1.00 | .35 | .41 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.96 | 0.89 | 0.83 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.89 | 0.83 | 0.94 | 0.55 | 0.39 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.95 | 0.56 | 0.40 | 0.48 | 0.32 | 0.24 | 0.00 | NaN | 0.00 |
| biblio weighted | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.60 | 0.54 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.45 | 0.40 | 0.36 | 0.90 | 0.53 | 0.37 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.98 | 0.57 | 0.40 | 0.48 | 0.30 | 0.22 | 0.00 | NaN | 0.00 |
| 1 | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.39 | 0.24 | NaN | NaN | 0.00 | 0.59 | 0.54 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.44 | 0.40 | 0.36 | 0.90 | 0.54 | 0.38 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.98 | 0.57 | 0.40 | 0.49 | 0.30 | 0.21 | 0.00 | NaN | 0.00 |
| 2 | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.39 | 0.24 | NaN | NaN | 0.00 | 0.60 | 0.54 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.45 | 0.40 | 0.36 | 0.90 | 0.53 | 0.37 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.98 | 0.57 | 0.40 | 0.48 | 0.29 | 0.21 | 0.00 | NaN | 0.00 |
| 3 | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.60 | 0.55 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.45 | 0.40 | 0.36 | 0.90 | 0.53 | 0.38 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.04 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.98 | 0.56 | 0.39 | 0.48 | 0.30 | 0.22 | 0.00 | NaN | 0.00 |
| 4 | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.60 | 0.55 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.45 | 0.40 | 0.36 | 0.91 | 0.52 | 0.37 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.98 | 0.57 | 0.40 | 0.49 | 0.31 | 0.23 | 0.00 | NaN | 0.00 |
| 5 | 1.00 | 1.00 | 1.00 | .58 | .54 | .50 | NaN | NaN | 0.00 | 1.00 | 0.38 | 0.24 | NaN | NaN | 0.00 | 0.60 | 0.54 | 0.50 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.45 | 0.40 | 0.36 | 0.91 | 0.52 | 0.36 | 0.43 | 0.46 | 0.50 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.98 | 0.57 | 0.40 | 0.48 | 0.31 | 0.22 | 0.00 | NaN | 0.00 |
| film | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | NaN | NaN | 0.00 | 1.00 | 0.32 | 0.20 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.81 | 0.70 | 0.83 | 0.13 | 0.07 | 0.62 | 0.51 | 0.44 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.78 | 0.60 | 0.49 | 0.59 | 0.07 | 0.03 | ||||||||||||
| 1 | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | NaN | NaN | 0.00 | 1.00 | 0.30 | 0.18 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.97 | 0.80 | 0.69 | 0.90 | 0.17 | 0.10 | 0.62 | 0.51 | 0.43 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.78 | 0.60 | 0.49 | 0.55 | 0.06 | 0.03 | ||||||||||||
| 2 | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | 1.00 | 0.31 | 0.18 | NaN | NaN | 0.00 | 0.97 | 0.80 | 0.69 | 0.75 | 0.06 | 0.03 | 0.62 | 0.51 | 0.44 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.78 | 0.60 | 0.49 | 0.56 | 0.04 | 0.02 | ||||||||||||||||||||||||||||||||||||
| 3 | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | 1.00 | 0.33 | 0.20 | NaN | NaN | 0.00 | 0.97 | 0.81 | 0.70 | 0.81 | 0.09 | 0.04 | 0.63 | 0.51 | 0.44 | 0.04 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.78 | 0.60 | 0.49 | 0.64 | 0.08 | 0.04 | ||||||||||||||||||||||||||||||||||||
| 4 | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | 1.00 | 0.33 | 0.20 | NaN | NaN | 0.00 | 0.97 | 0.81 | 0.70 | 0.87 | 0.19 | 0.11 | 0.62 | 0.51 | 0.44 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.78 | 0.60 | 0.49 | 0.60 | 0.08 | 0.04 | ||||||||||||||||||||||||||||||||||||
| 5 | 1.00 | 1.00 | 1.00 | 0.43 | 0.47 | 0.50 | 1.00 | 0.35 | 0.21 | NaN | NaN | 0.00 | 0.97 | 0.81 | 0.70 | 0.85 | 0.13 | 0.07 | 0.62 | 0.51 | 0.43 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.77 | 0.60 | 0.49 | 0.62 | 0.08 | 0.04 | ||||||||||||||||||||||||||||||||||||
| H-mean | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | NaN | NaN | 0.00 | 0.99 | 0.32 | 0.20 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.79 | 0.12 | 0.06 | 0.62 | 0.51 | 0.44 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.62 | 0.49 | 0.58 | 0.06 | 0.03 | ||||||||||||
| 1 | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | NaN | NaN | 0.00 | 0.99 | 0.30 | 0.18 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.83 | 0.16 | 0.09 | 0.62 | 0.51 | 0.43 | NaN | NaN | 0.00 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.62 | 0.49 | 0.56 | 0.05 | 0.03 | ||||||||||||
| 2 | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | 0.99 | 0.31 | 0.18 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.72 | 0.06 | 0.03 | 0.62 | 0.51 | 0.44 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.62 | 0.49 | 0.57 | 0.03 | 0.02 | ||||||||||||||||||||||||||||||||||||
| 3 | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | 0.99 | 0.33 | 0.20 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.77 | 0.08 | 0.04 | 0.63 | 0.51 | 0.44 | 0.04 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.62 | 0.49 | 0.56 | 0.06 | 0.03 | ||||||||||||||||||||||||||||||||||||
| 4 | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | 0.99 | 0.33 | 0.20 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.83 | 0.18 | 0.10 | 0.62 | 0.51 | 0.44 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.62 | 0.49 | 0.59 | 0.07 | 0.04 | ||||||||||||||||||||||||||||||||||||
| 5 | 1.00 | 1.00 | 1.00 | 0.68 | 0.58 | 0.50 | 0.99 | 0.35 | 0.21 | NaN | NaN | 0.00 | 0.39 | 0.31 | 0.26 | 0.82 | 0.12 | 0.07 | 0.62 | 0.51 | 0.43 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.01 | 0.84 | 0.61 | 0.49 | 0.63 | 0.07 | 0.04 | ||||||||||||||||||||||||||||||||||||
n/a: result alignment not provided or not readable
NaN: division per zero, likely due to empty alignment.
The full set of resulting alignments can be found at biblio.zip and film.zip.
This track has been performed by Jérôme Euzenat. If you have any problems working with the ontologies, any questions or suggestions, feel free to contact him.
[1] Jérôme Euzenat, Maria Roşoiu, Cássia Trojahn dos Santos. Ontology matching benchmarks: generation, stability, and discriminability, Journal of web semantics 21:30-48, 2013 [DOI:10.1016/j.websem.2013.05.002]