

The output alignments can be downloaded here.
The main focus of the evaluation here is to be task-oriented. First, we manually evaluate the quality of the generated alignments, in terms of precision. Second, we evaluate the generated correspondences using a SPARQL query rewriting system and manually measure their ability of answering a set of queries over each dataset.
We excluded from the evaluation the generated correspondences implying identical IRIs such as owl:Class = owl:Class. As instance matching is not the focus of this evaluation, we do not consider instance correspondences. In order to measure the Precision of the output alignments, each correspondence (except if in the categories mentioned above) was manually verified and classified:
This gives 4 Precision scores:
The confidence of the correspondence is not taken into account. All the systems output only equivalence correspondences so the relation of the correspondence was not considered in the evaluation.
6 reference equivalent queries per ontology are used to assess how an alignment covers them. The alignment is used to rewrite every reference. For each reference query, the best rewritten query is classified as equivalent, more specific, more general or overlapping.
The same scores as for the Precision are calculated for the Coverage:
All the other systems (AGM, AML, CANARD, FCAMap-KG, LogMap, LogMapBio, LogMapLt, ONTMAT1, POMAP++) output at least an alignment. ONTMAT1 only output correspondences with identical URIs which were filtered for the evaluation. The only system to output complex correspondences was CANARD.
This year, every system was evaluated on their oriented alignment output, especially for the Coverage calculation. For example, let's take a set of equivalent correspondences: Q={SELECT ?x WHERE{ ?x a agtx:Taxon}, SELECT ?x WHERE{ ?x a dbo:Species}}. If an output alignment agronomicTaxon-dbpedia contains (agtx:Taxon,dbo:Species,≡) but the alignment dbpedia-agronomicTaxon does NOT contain (dbo:Species, agtx:Taxon, ≡). The coverage score of Q for the pair agronomicTaxon-dbpedia is 1 but the coverage score of Q for dbpedia-agronomicTaxon is 0. Last year the evaluation was non-oriented, so the coverage score of Q would be the same (1.0) for agronomicTaxon-dbpedia and dbpedia-agronomicTaxon.
The systems have been executed on a Ubuntu 16.04 machine configured with 16GB of RAM running under a i7-4790K CPU 4.00GHz x 8 processors. All measurements are based on a single run.
Number of correspondences per type per system and runtime over the track.
"identical" shows the number of correspondences which align two identical URIs, instance, the number of correspondences which align two instances
| tool | (1:1) | (1:n) | (m:1) | (m:n) | identical | instance | total | filtered | runtime(s) | runtime (min) |
|---|---|---|---|---|---|---|---|---|---|---|
| AGM | 169 | 0 | 0 | 0 | 11 | 3029 | 3209 | 169 | 11593 | 193 |
| AML | 30 | 0 | 0 | 0 | 0 | 0 | 30 | 30 | 2444 | 41 |
| CANARD | 67 | 540 | 15 | 169 | 0 | 0 | 791 | 791 | 30712 | 512 |
| FCAMap-KG | 20 | 0 | 0 | 0 | 10 | 0 | 30 | 20 | 1284 | 21 |
| LogMap | 38 | 0 | 0 | 0 | 58 | 0 | 96 | 38 | 1544 | 26 |
| LogMapBio | 36 | 0 | 0 | 0 | 54 | 0 | 90 | 36 | 2009 | 33 |
| LogMapKG | 34 | 0 | 0 | 0 | 58 | 12776 | 12868 | 34 | 1654 | 28 |
| LogMapLt | 972 | 0 | 0 | 0 | 1528 | 7628 | 10128 | 972 | 1503 | 25 |
| ONTMAT1 | 0 | 0 | 0 | 0 | 12 | 0 | 12 | 0 | 1586 | 26 |
| POMAP++ | 1 | 0 | 0 | 0 | 25 | 7 | 33 | 1 | 2252 | 38 |
Results over the Taxon dataset
| Precision | Coverage |
|||||||
|---|---|---|---|---|---|---|---|---|
| tool | classical | recall oriented | precision oriented | overlap | classical | recall oriented | precision oriented | overlap |
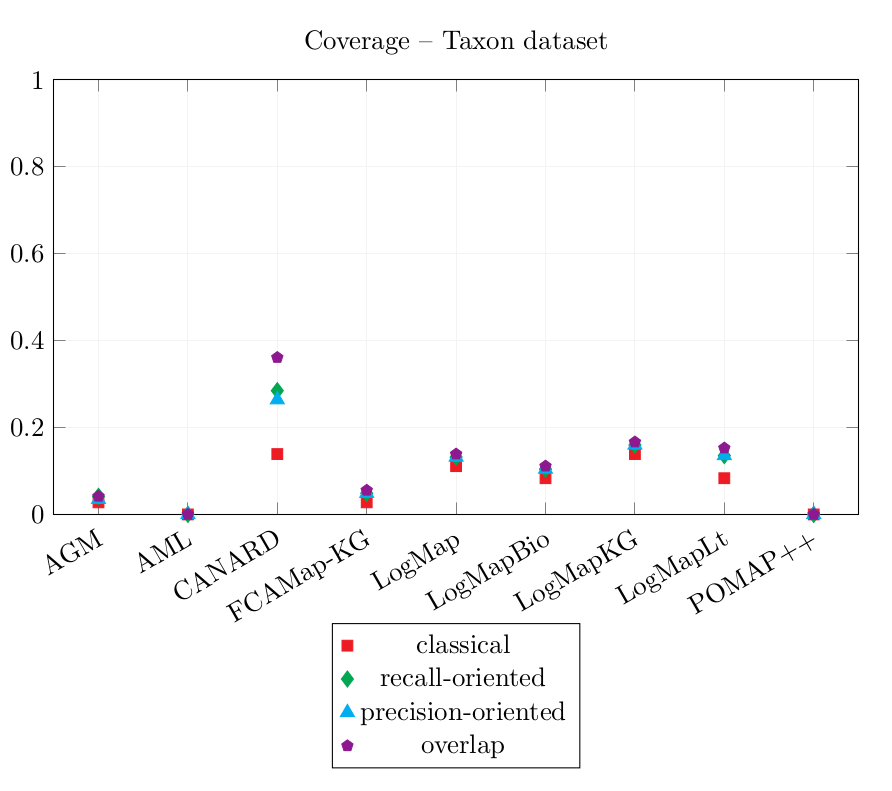
| AGM | 0.055 | 0.122 | 0.101 | 0.134 | 0.028 | 0.042 | 0.035 | 0.042 |
| AML | 0.533 | 0.533 | 0.533 | 0.533 | 0 | 0 | 0 | 0 |
| CANARD | 0.083 | 0.455 | 0.363 | 0.914 | 0.139 | 0.285 | 0.264 | 0.361 |
| FCAMap-KG | 0.625 | 0.875 | 0.875 | 0.958 | 0.028 | 0.049 | 0.049 | 0.056 |
| LogMap | 0.625 | 0.729 | 0.729 | 0.792 | 0.111 | 0.132 | 0.132 | 0.139 |
| LogMapBio | 0.537 | 0.659 | 0.659 | 0.723 | 0.083 | 0.104 | 0.104 | 0.111 |
| LogMapKG | 0.547 | 0.641 | 0.641 | 0.686 | 0.139 | 0.16 | 0.16 | 0.167 |
| LogMapLt | 0.221 | 0.365 | 0.365 | 0.413 | 0.083 | 0.135 | 0.135 | 0.153 |
| POMAP++ | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
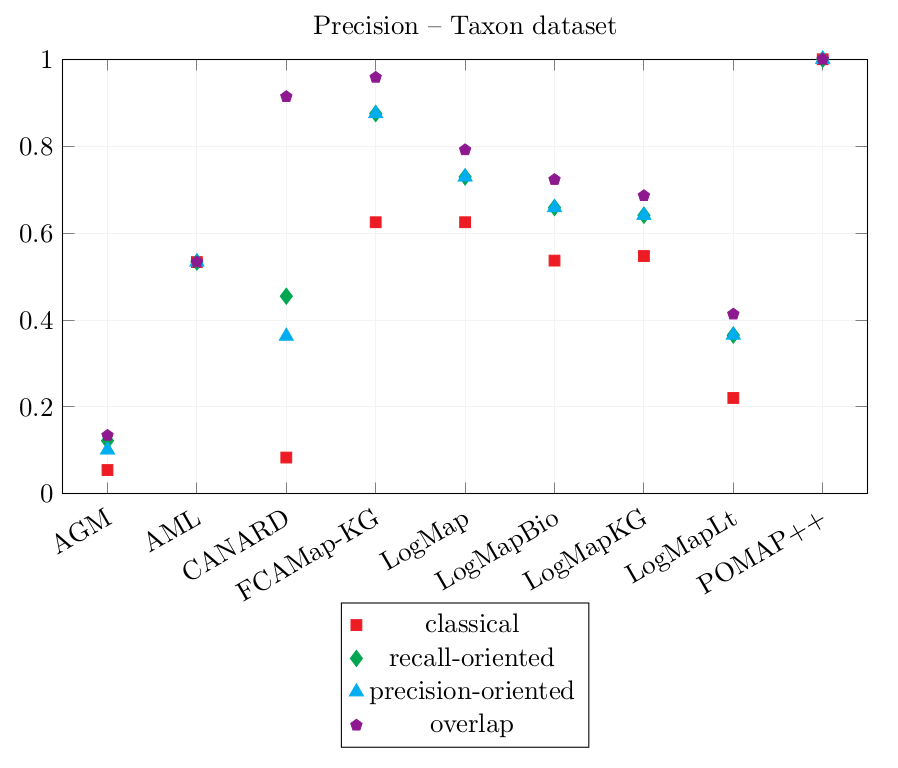
LogMapLt and AGM have output correspondences such as agronomicTaxon:PhylumRank = dbo:phylum. As the source member is a class and the target member is an object property, these correspondences were counted as incorrect. However, they would be equivalent if transformed into complex correspondences such as (agronomicTaxon:PhylumRank, Exists dbo:phylum^-, ≡).
CANARD has the longest runtime by far. It took 8h32min to complete the Taxon matching task. It is the only matcher which outputs complex correspondences on this dataset.
The two system which cover the most queries by enabling an equivalent query output are LogMapKG and CANARD: their classical Coverage score is 0.139. CANARD enables to cover more queries with more specific, more general or overlapping answers.
In comparison with last year’s results, the Coverage score of CANARD, LogMap and LogMapBio have slightly improved (comparing only equivalent queries). This shows improvement especially in the case of oriented aligning systems such as CANARD as last year's evaluation was not oriented.
The classical Precision score of CANARD has decreased in comparison with last year's results while those of AML, LogMap, LogMapBio, LogMapLt, POMAP++ have increased.
POMAP++ output only one correspondence which was not filtered. This correspondence is correct, therefore, POMAP++ has the highest Precision scores 1.0. The correspondence could not be used to rewrite the reference queries, so its Coverage scores are 0.0.
With respect to the technical environment, the SEALS system as it is now was probably not adapted to deal with big knowledge bases as the loading phase got very slow. The use of SPARQL endpoints instead of two ontology files would make more sense given that many LOD repositories provide one.