

Complex alignments are more expressive than simple alignments as their correspondences can contain logical constructors or transformation functions of literal values.
For example, given two ontologies o1 and o2:With this track, we evaluate systems which can generate such correspondences.
The complex track contains eight datasets from various domain: Conference, Populated Conference, Hydrography, GeoLink, Populated GeoLink, Populated Enslaved, and Biomedical. Each of the datasets and corresponding evaluation methods are presented below.
The participants of the track should output their (complex) correspondences in the EDOAL format. This format is supported by the Alignment API. The evaluation will be supported by the MELT framwork this year. The participants have to wrap their tool as described at MELT framework. For executing the tasks in each dataset the parameters are listed in boxes below (repository, suite-id, version-id).
The number of ontologies, simple (1:1) and complex (1:n), (m:n) correspondences for each dataset of this track are summarized in the following table.
| Dataset | #Ontologies | #(1:1) | #(1:n) | #(m:n) |
|---|---|---|---|---|
| Conference | 3 | 78 | 79 | 0 |
| Populated Conference | 5 | 111 | 86 | 98 |
| Hydrography | 4 | 113 | 69 | 15 |
| GeoLink | 2 | 19 | 5 | 43 |
| Populated GeoLink | 2 | 19 | 5 | 43 |
| Populated Enslaved | 2 | 15 | 0 | 83 |
| Taxon | 4 | 6 | 17 | 3 |
| Biomedical | 17 | - | 15627 | 0 |
This dataset is based on the OntoFarm dataset [1] used in the Conference track of the OAEI campaigns. It is composed of 16 ontologies on the conference organisation domain and simple reference alignments between 7 of them. Here, we consider 3 out of the 7 ontologies from the reference alignments (cmt, conference and ekaw), resulting in 3 alignment pairs.
The correspondences were manually curated by 3 experts following the query rewriting methodology in [2]. For each pair o1-o2 of ontologies, the following steps were applied:
4 experts assessed the curated correspondences to reach a consensus.
In order to allow matchers which rely on instances to participate over the Conference complex track, we propose a populated version of the Conference dataset. 5 ontologies have been populated with more or less common instances resulting in 6 datasets: (6 versions on the repository: v0, v20, v40, v60, v80 and v100).
The hydrography dataset is composed of four source ontologies, which are Hydro3, HydrOntology_native, HydrOntology_translated, and Cree, that each should be aligned to a single target Surface Water Ontology (SWO). The source ontologies vary in their similarity to the target ontology -- Hydro3 is similar in both language and structure, hydrOntology is similar in structure but is in Spanish rather than English, and Cree is very different in terms of both language and structure.
The alignments were created by a geologist and an ontologist, in consultation with a native Spanish speaker regarding the hydrOntology, and consist of logical relations.
There are three subtasks in the Hydrography complex alignment track:
For each entity in the source ontology, the alignment system is asked to list all of the entities in the target ontologies that are related to it in some way.
For example:
owl:equivalentClasses(ont1:A1 owl:intersectionOf(ont2:B1 owl:someValuesFrom(ont2:B2 ont2:B3))
The goal in this task is to find the most relevant entities in the ont2 to the class ont1:A1. In this case, the best output would be ont2:B1, ont2:B2, and ont2:B3.
For each alignmnet, the system should then endeavor to find the concrete relationships, such as equivalence, subsumption, intersection, value restriction, and so on, that hold between the entities. In terms of the example above, an alignment system needs to eventually determine that the relationship between the two sides is equivalence.
This task is a combination of the two former steps.
This dataset is from the GeoLink project, which was funded under the U.S. National Science Foundation's EarthCube initiative. It is composed of two ontologies: the GeoLink Base Ontology (GBO) and the GeoLink Modular Ontology (GMO). The GeoLink project is a real-world use case of ontologies, and its instance data is available. The alignment between the two ontologies was developed in consultation with domain experts from several geoscience research institutions. This alignment is a slightly simplified version of the one discussed in [4]. The relations that involve punning have been removed due to a concern that many automated alignment systems would not consider these as potential mappings. More details can be found in [4].
In order to allow alignment systems that rely on the instance data to participate over the Geolink complex track, we also generate a populated version of the Geolink dataset. The instance data are from real-worlds and collected from seven data repositories in the Geolink project. More details of the populated geolink dataset can be found in [8].
This dataset is from the Enslaved project, which was funded under The Andrew w. Mellon Foundation. It is composed of two resources: the Enslaved Ontology and the Enslaved Wikidata Knowledge Graph. The Enslaved project is a real-world use case of ontologies in pepple of historic slave trade domain, and its instance data is available and already incorporated into Wikidata repository. The alignment between the two resources was developed in consultation with domain experts from several historian research institutions. More details of the populated enslaved dataset can be found in [9].
This dataset was constructed from the existing logical definitions in three biomedical ontologies: Human Phenotype ontology (HP), Mammalian Phenotype ontology (MP), and Worm Phenotype ontology (WBP). Logical definitions are 1:n complex mappings that use multiple target ontologies in their construction and therefore this task serves as a complex multi-ontology matching (CMOM) challenge.
Both HP and MP have the same set of target ontologies: Cell ontology (CL), Chemical Entities of Biological Interest (ChEBI), Gene Ontology (GO), Phenotype and Trait Ontology (PATO), and Uber Anatomy Ontology (UBERON). WBP uses ChEBI, GO, PATO, and the C.elegans Gross Anatomy Ontology (WBbt).
The reference constructs can be found at https://dx.doi.org/10.21227/btb7-yd20 [13].
The complex correspondences output by the systems will be manually compared to the ones of the consensus alignment.
For this first evaluation, only equivalence correspondences will be evaluated and the confidence of the correspondenes will not be taken into account.
The systems can take the ra1 simple alignments as input. The ra1 alignments can be downloaded here.
Complex constructs were evaluated based on whether they use the expected correct classes from the reference mapping and on the Graph Edit Distance (GED) necessary to transform the generated mapping into the reference mapping.
Class correction:
Partial correctness was assessed by extracting the classes used in the generated mapping and comparing them to the reference classes, following an adaptation of the class evaluation described in [8]. Each mapping is sorted into one of the following categories: i) correct, all classes match, ii) contained in reference, generated classes are completely contained within the set of reference classes, iii) contains reference, reference classes are completely contained within the set of generated classes, iv) overlap, some classes are shared between the sets, and v) incorrect, there are no shared classes in between the two sets. This metric evaluates all mappings: simple 1:1, complex 1:n, and complex n:m.
Graph Edit Distance (GED):
To assess both the class selection and their combination into a complex construct, each mapping is converted into a graph (an example is shown in Figure 1) and the graph edit distance from generated mapping to reference mapping is computed. This is performed using the NetworkX package [9] and follows the equations and strategies outlined in [10]. This metric evaluates simple 1:1 mappings and complex 1:n mappings.
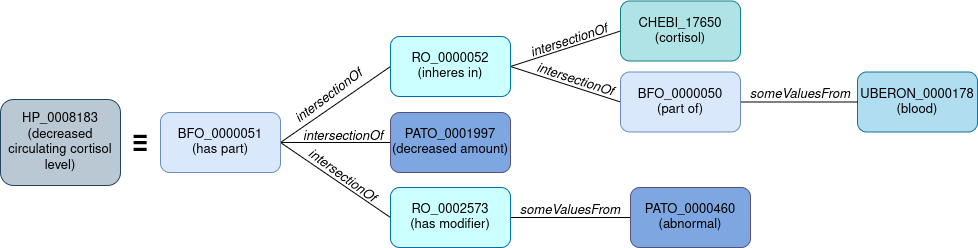
Figure 1 - Graph conversion of the logical definition HP_0008183 “decreased circulating cortisol level” from the Human Phenotype ontology [11].
Tree Edit Distance (TED)
The other evaluation metric used is proposed in ref[12]. This metric compares the matcher proposed alignment with the reference alignment using Tree Edit Distance (TED) and finds the most similar pairs of correspondences using an assignment algorithm. The metric also follows desired properties for complex matching evaluation based on reference alignments.
[1] Ondřej Zamazal, Vojtěch Svátek. The Ten-Year OntoFarm and its Fertilization within the Onto-Sphere. Web Semantics: Science, Services and Agents on the World Wide Web, 43, 46-53. 2017.
[2] Élodie Thiéblin, Ollivier Haemmerlé, Nathalie Hernandez, Cassia Trojahn. Task-Oriented Complex Ontology Alignment: Two Alignment Evaluation Sets. In : European Semantic Web Conference. Springer, Cham, 655-670, 2018.
[3] Élodie Thiéblin, Fabien Amarger, Nathalie Hernandez, Catherine Roussey, Cassia Trojahn. Cross-querying LOD datasets using complex alignments: an application to agronomic taxa. In: Research Conference on Metadata and Semantics Research. Springer, Cham, 25-37, 2017.
[4] Lu Zhou, Michelle Cheatham, Adila Krisnadhi, Pascal Hitzler. A Complex Alignment Benchamark: GeoLink Dataset. In: International Semantic Web Conference. Springer, Proceedings, Part II, pp. 273-288, 2018.
[5] Marc Ehrig, and Jérôme Euzenat. "Relaxed precision and recall for ontology matching." K-CAP 2005 Workshop on Integrating Ontologies, Banff, Canada, 2005.
[6] Élodie Thiéblin. Do competency questions for alignment help fostering complex correspondences?. In EKAW Doctoral Consortium, 2018.
[7] Élodie Thiéblin, Fabien Amarger, Ollivier Haemmerlé, Nathalie Hernandez, Cassia Trojahn. Rewriting SELECT SPARQL queries from 1:n complex correspondences. In: Ontology Matching, pp. 49-60, 2016.
[8] Silva, M. C., Faria, D., & Pesquita, C. (2024). Complex Multi-Ontology Alignment Through Geometric Operations on Language Embeddings. In ECAI 2024 (pp. 1333–1340). IOS Press. https://doi.org/10.3233/FAIA240632
[9] Aric A. Hagberg, Daniel A. Schult and Pieter J. Swart, “Exploring network structure, dynamics, and function using NetworkX”, in Proceedings of the 7th Python in Science Conference (SciPy2008), Gäel Varoquaux, Travis Vaught, and Jarrod Millman (Eds), (Pasadena, CA USA), pp. 11–15, Aug 2008
[10] Silva, M. C., Faria, D., & Pesquita, C. (2025). CMOMgen: Complex Multi-Ontology Alignment via Pattern-Guided In-Context Learning (No. arXiv:2510.21656). arXiv. https://doi.org/10.48550/arXiv.2510.21656
[11] Gargano MA et al., The Human Phenotype Ontology in 2024: phenotypes around the world. Nucleic Acids Res. 2024 Jan 5;52(D1):D1333-D1346. doi: 10.1093/nar/gkad1005. PMID: 37953324; PMCID: PMC10767975., https://doi.org/10.1093/nar/gkad1005
[12] Santos Sousa, G., Lima, R., Trojahn, C. (2025). On Evaluation Metrics for Complex Matching Based on Reference Alignments. In: Curry, E., et al. The Semantic Web. ESWC 2025. Lecture Notes in Computer Science, vol 15718. Springer, Cham. https://doi.org/10.1007/978-3-031-94575-5_5
[13] Marta Silva, Daniel Faria, Catia Pesquita, "Reference Alignments for biomedical Complex Multi-Ontology Matching tasks (HP, MP, WBP)", IEEE Dataport, September 3, 2025, doi:10.21227/btb7-yd20