

In the following, are the results of the OAEI 2014 evaluation of the benchmarks track.
All tests have been run entirely from the SEALS platform with the strict same protocol.
This year we run a campaign with three new generated tests. We did not run this year scalability tests.
Evaluations were run on a Debian Linux virtual machine configured with four processors and 8GB of RAM running under a Dell PowerEdge T610 with 2*Intel Xeon Quad Core 2.26GHz E5607 processors and 32GB of RAM, under Linux ProxMox 2 (Debian).
All matchers where run under the SEALS client using Java 1.7 and a maximum heap size of 8GB (which has been necessary for the larger tests, i.e., dog). No timeout was explicitly set.
Reported figures are the average of 5 runs. As has already been shown in [1], there is not much variance in compliance measures across runs. This is not necessarily the case for time measurements so we report standard deviations with time measurements.
From the 13 systems listed in the 2014 final results page, 10 systems participated in this track. A few of these systems encountered problems:
The following table presents the harmonic means of precision, F-measure and recall for the test suites for all the participants, along with their confidence-weighted values. The table also shows measures provided by edna, a simple edit distance algorithm on labels which is used as a baseline.
| biblio | cose | dog | |||||||
| Matching system | Prec. | F-m. | Rec. | Prec. | F-m. | Rec. | Prec. | F-m. | Rec. |
| edna | .35(.58) | .41(.54) | .50 | .44(.72) | .47(.59) | .50 | .50(.74) | .50(.60) | .50 |
| AML | .92(.94) | .55(.56) | .39 | .46(.59) | .46(.51) | .46(.45) | .98(.96) | .73(.71) | .58(.57) |
| AOT_2014 | .80(.90) | .64(.67) | .53 | .69(.84) | .58(.63) | .50 | .62(.77) | .62(.68) | .61 |
| AOTL | .85(.89) | .65(.66) | .53 | .94(.95) | .65(.65) | .50 | .97 | .74(.75) | .60 |
| LogMap | .40(.40) | .40(.39) | .40(.37) | .38(.45) | .41(.40) | .45(.37) | .96(.91) | .15(.14) | .08(.07) |
| LogMap-C | .42(.41) | .41(.39) | .40(.37) | .39(.45) | .41(.40) | .43(.35) | .98(.92) | .15(.13) | .08(.07) |
| LogMapLite | .43 | .46 | .50 | .37 | .43 | .50 | .86 | .71 | .61 |
| MaasMtch | .97 | .56 | .39 | .98 | .48 | .31 | .92 | .55 | .39 |
| OMReasoner | .73 | .59 | .50 | .08 | .14 | .50 | * | * | * |
| RSDLWB | .99 | .66 | .50 | * | * | * | .99 | .75 | .60 |
| XMap2 | 1.0 | .57 | .40 | 1.0 | .28 | .17 | 1.0 | .32 | .20 |
Some system have had constant problems with the most strongly altered tests to the point of not outputing results: LogMap-C, LogMap, MaasMtch. Problems were also encountered to a smaller extent by XMap2. OMReasoner failed to return any answer on dog, and RSDLWB on cose.
Concerning F-measure results, the AOTL system seems to achieve the best results before RSDLWB. AOTL is also well balanced: it always achieve more than 50% recall with still a quite high precision. RSDLWD is slightly better than AOTL on two tests but did not provide results on the third one.
AOT_2014 is a close follower of AOTL. AML had very good results on dog and OMReasoner on biblio.
In general, results of the best matchers are largely lower than those of the best matchers in the previous year.
We can consider that we have high-precision matchers (XMap2: 1.0, RSDLWB: .99, MaasMatch: .92-.98; AML: (.46)-.98). LogMap-C, LogMap achieve also very high precision in dog (their other bad precision are certainly due to LogMap matching enties out of the ontology namespaces). Of these high-precision matchers, RSDLWB is remarkable since it achieves a 50% recall (when it works).
The recall of systems is generally high with figures around 50% but this may be due to the structure of benchmarks.
Confidence-weighted measures reward systems able to provide accurate confidence values. Using confidence-weighted F-measures usually increase F-measure of systems showing that thay are able to provide a meaningfull assessment of their correspondences. The exception to this rule is LogMap whose weighted values are lower. Again, this may be due to the out put of correspondences out of the ontology namespace or instance correspondences.
| biblio | cose | dog | |||||||
| Matching system | s. | stdev | F-m./s. | s. | stdev | F-m./s. | s. | stdev | F-m./s. |
| AML | 48.96 | 0.59 | 1.12 | 140.29 | 1.37 | 0.33 | 1506.16 | 81.66 | 0.05 |
| AOT_2014 | 166.91 | 1.85 | 0.38 | 194.02 | 1.31 | 0.30 | 10638.27 | 82.37 | 0.01 |
| AOTL | 741.98 | 8.37 | 0.09 | 386.18 | 7.50 | 0.17 | 18618.60 | 268.90 | 0.00 |
| LogMap | 106.68 | 0.90 | 0.37 | 123.44 | 1.79 | 0.33 | 472.31 | 74.00 | 0.03 |
| LogMap-C | 158.36 | 0.84 | 0.26 | 188.30 | 2.29 | 0.22 | 953.56 | 180.58 | 0.02 |
| LogMapLite | 61.43 | 0.65 | 0.75 | 62.67 | 0.93 | 0.69 | 370.32 | 90.76 | 0.19 |
| MaasMtch | 122.50 | 2.75 | 0.46 | 392.43 | 6.97 | 0.12 | 7338.92 | 135.61 | 0.01 |
| OMReasoner | 60.01 | 0.26 | 0.98 | 98.17 | 0.89 | 0.14 | 331.65 | 196.85 | * |
| RSDLWB | 86.22 | 1.75 | 0.77 | * | * | * | 14417.32 | 285.51 | 0.01 |
| XMap2 | 68.67 | 0.65 | 0.83 | 31.39 | 12.24 | 0.89 | 221.83 | 122.99 | 0.14 |
We provide the average time, times standard deviation and F-measure point provided per second by matchers. Standard deviation should be better reported in percentage of the total time. The F-measure point provided per second shows that efficient matchers are XMap2 and LogMapLite followed by AML (these results are consistent on cose and dog, biblio is a bit different but certainly due to errors reported above).
We draw the triangle graphs for the three tests. It confirms the impressions above: systems are very precision-oriented but AOT_2014 which stands in the middle of the graph. AOTL has, in general, good results.
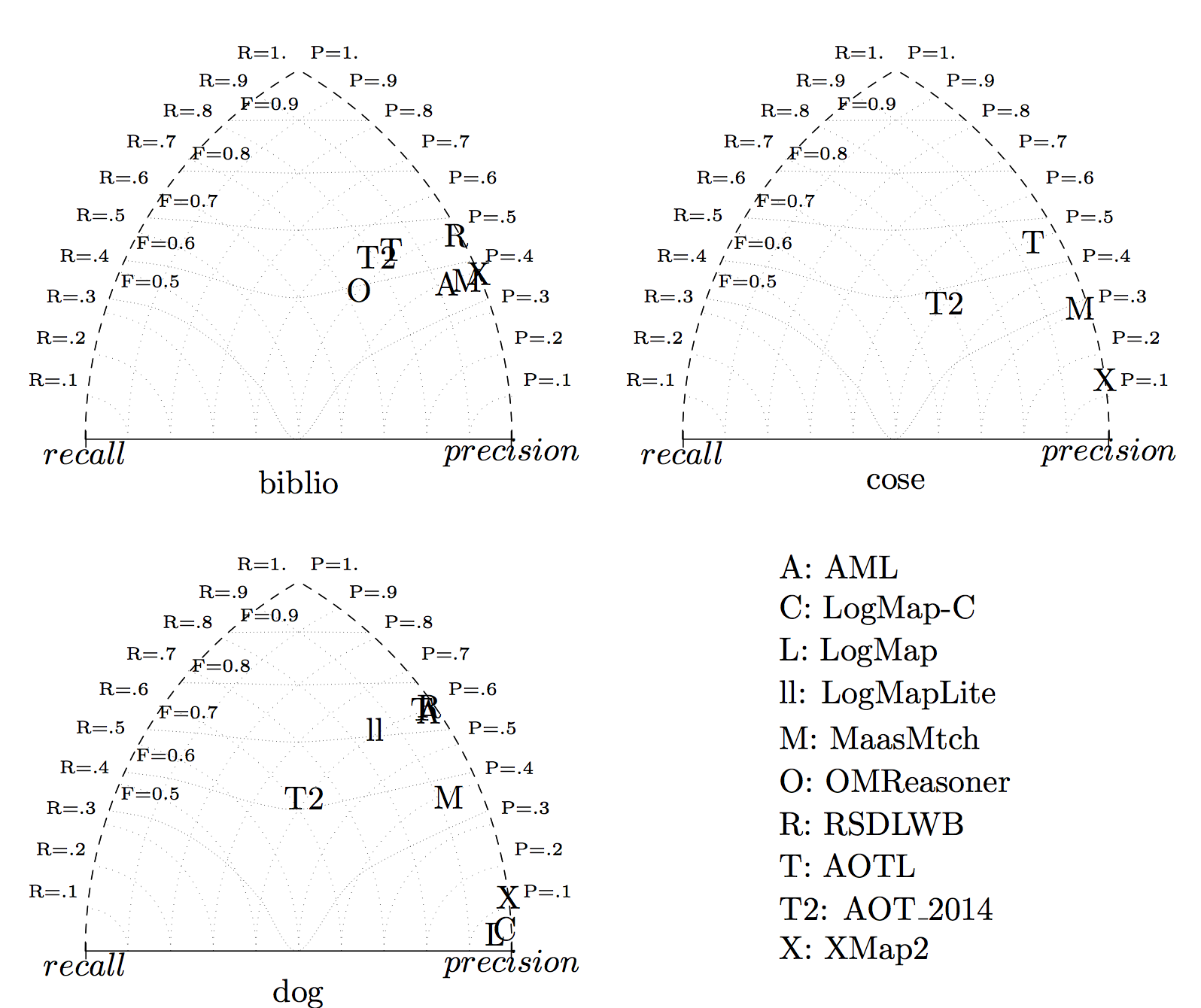
You can obtain the detailed results, tests by tests, by clicking on the test run number.
| algo | refalign | edna | AOT_2014 | AOTL | LogMapLite | LogMap-C | LogMap | MaasMtch | AML | XMap2 | RSDLWB | OMReasoner | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| test | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. | Prec. | FMeas. | Rec. |
| biblio | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.80 | 0.64 | 0.53 | 0.85 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.42 | 0.41 | 0.40 | 0.40 | 0.40 | 0.40 | 0.97 | 0.56 | 0.39 | 0.92 | 0.55 | 0.39 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 1 | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.81 | 0.64 | 0.53 | 0.85 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.42 | 0.41 | 0.40 | 0.40 | 0.40 | 0.40 | 0.97 | 0.56 | 0.39 | 0.91 | 0.54 | 0.38 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 2 | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.81 | 0.64 | 0.53 | 0.85 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.41 | 0.40 | 0.40 | 0.39 | 0.39 | 0.40 | 0.97 | 0.56 | 0.39 | 0.92 | 0.55 | 0.40 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 3 | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.80 | 0.64 | 0.53 | 0.84 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.43 | 0.41 | 0.39 | 0.41 | 0.40 | 0.39 | 0.97 | 0.56 | 0.39 | 0.92 | 0.56 | 0.40 | 1.00 | 0.58 | 0.41 | 0.99 | 0.66 | 0.49 | 0.73 | 0.59 | 0.50 |
| 4 | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.80 | 0.64 | 0.53 | 0.85 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.42 | 0.41 | 0.39 | 0.40 | 0.40 | 0.39 | 0.97 | 0.55 | 0.39 | 0.92 | 0.55 | 0.39 | 1.00 | 0.56 | 0.39 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 5 | 1.00 | 1.00 | 1.00 | 0.35 | 0.41 | 0.50 | 0.80 | 0.64 | 0.53 | 0.84 | 0.65 | 0.53 | 0.43 | 0.46 | 0.50 | 0.43 | 0.42 | 0.41 | 0.41 | 0.41 | 0.41 | 0.97 | 0.55 | 0.39 | 0.92 | 0.55 | 0.39 | 1.00 | 0.57 | 0.39 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| biblio: weighted | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.90 | 0.67 | 0.53 | 0.89 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.41 | 0.39 | 0.37 | 0.40 | 0.39 | 0.37 | 0.97 | 0.56 | 0.39 | 0.94 | 0.56 | 0.39 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 1 | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.90 | 0.67 | 0.53 | 0.89 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.41 | 0.39 | 0.38 | 0.40 | 0.39 | 0.38 | 0.97 | 0.56 | 0.39 | 0.94 | 0.55 | 0.38 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 2 | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.90 | 0.67 | 0.53 | 0.90 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.40 | 0.39 | 0.37 | 0.40 | 0.38 | 0.37 | 0.97 | 0.56 | 0.39 | 0.94 | 0.56 | 0.40 | 1.00 | 0.57 | 0.40 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 3 | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.89 | 0.67 | 0.53 | 0.88 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.42 | 0.39 | 0.36 | 0.41 | 0.39 | 0.36 | 0.98 | 0.56 | 0.39 | 0.94 | 0.56 | 0.40 | 1.00 | 0.58 | 0.41 | 0.99 | 0.66 | 0.49 | 0.73 | 0.59 | 0.50 |
| 4 | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.90 | 0.67 | 0.53 | 0.89 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.41 | 0.39 | 0.37 | 0.41 | 0.39 | 0.37 | 0.98 | 0.56 | 0.39 | 0.94 | 0.56 | 0.39 | 1.00 | 0.56 | 0.39 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| 5 | 1.00 | 1.00 | 1.00 | 0.58 | 0.54 | 0.50 | 0.90 | 0.67 | 0.53 | 0.89 | 0.66 | 0.53 | 0.43 | 0.46 | 0.50 | 0.41 | 0.40 | 0.39 | 0.40 | 0.40 | 0.39 | 0.97 | 0.55 | 0.39 | 0.94 | 0.56 | 0.39 | 1.00 | 0.57 | 0.39 | 0.99 | 0.66 | 0.50 | 0.73 | 0.59 | 0.50 |
| cose | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.39 | 0.41 | 0.43 | 0.38 | 0.41 | 0.45 | 0.98 | 0.48 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.28 | 0.17 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 1 | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.38 | 0.40 | 0.42 | 0.38 | 0.42 | 0.47 | 0.98 | 0.48 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.27 | 0.15 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 2 | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.39 | 0.42 | 0.45 | 0.38 | 0.41 | 0.45 | 0.98 | 0.47 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.26 | 0.15 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 3 | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.38 | 0.40 | 0.42 | 0.38 | 0.40 | 0.43 | 0.98 | 0.48 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.45 | 0.29 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 4 | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.39 | 0.42 | 0.46 | 0.38 | 0.42 | 0.47 | 0.98 | 0.48 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.19 | 0.10 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 5 | 1.00 | 1.00 | 1.00 | 0.44 | 0.47 | 0.50 | 0.69 | 0.58 | 0.50 | 0.94 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.39 | 0.40 | 0.42 | 0.38 | 0.40 | 0.43 | 0.98 | 0.47 | 0.31 | 0.46 | 0.46 | 0.46 | 1.00 | 0.24 | 0.14 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| cose: weighted | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.95 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.45 | 0.40 | 0.35 | 0.45 | 0.40 | 0.37 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.45 | 1.00 | 0.28 | 0.17 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 1 | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.95 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.45 | 0.39 | 0.34 | 0.45 | 0.41 | 0.38 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.45 | 1.00 | 0.27 | 0.15 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 2 | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.95 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.45 | 0.40 | 0.36 | 0.44 | 0.40 | 0.37 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.45 | 1.00 | 0.26 | 0.15 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 3 | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.95 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.44 | 0.38 | 0.34 | 0.45 | 0.39 | 0.35 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.45 | 1.00 | 0.45 | 0.29 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 4 | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.95 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.44 | 0.40 | 0.37 | 0.44 | 0.41 | 0.38 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.46 | 1.00 | 0.19 | 0.10 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| 5 | 1.00 | 1.00 | 1.00 | 0.72 | 0.59 | 0.50 | 0.84 | 0.63 | 0.50 | 0.96 | 0.65 | 0.50 | 0.37 | 0.43 | 0.50 | 0.45 | 0.39 | 0.34 | 0.45 | 0.39 | 0.35 | 0.98 | 0.48 | 0.31 | 0.59 | 0.51 | 0.45 | 1.00 | 0.24 | 0.14 | NaN | NaN | 0.00 | 0.08 | 0.14 | 0.50 |
| dog | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.62 | 0.62 | 0.61 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.98 | 0.15 | 0.08 | 0.96 | 0.15 | 0.08 | 0.92 | 0.55 | 0.39 | 0.98 | 0.73 | 0.58 | 1.00 | 0.32 | 0.20 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 1 | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.62 | 0.62 | 0.61 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.98 | 0.18 | 0.10 | 0.96 | 0.19 | 0.10 | 0.93 | 0.54 | 0.39 | 0.98 | 0.74 | 0.59 | 1.00 | 0.30 | 0.17 | 0.99 | 0.75 | 0.61 | NaN | NaN | 0.00 |
| 2 | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.62 | 0.62 | 0.61 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.98 | 0.10 | 0.05 | 0.96 | 0.10 | 0.05 | 0.92 | 0.55 | 0.39 | 0.98 | 0.73 | 0.58 | 1.00 | 0.41 | 0.26 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 3 | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.63 | 0.62 | 0.61 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.99 | 0.15 | 0.08 | 0.96 | 0.15 | 0.08 | 0.92 | 0.55 | 0.39 | 0.98 | 0.74 | 0.59 | 1.00 | 0.22 | 0.13 | 0.99 | 0.75 | 0.61 | NaN | NaN | 0.00 |
| 4 | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.62 | 0.61 | 0.60 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.98 | 0.20 | 0.11 | 0.96 | 0.20 | 0.11 | 0.93 | 0.55 | 0.39 | 0.98 | 0.72 | 0.56 | 1.00 | 0.45 | 0.29 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 5 | 1.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 0.62 | 0.61 | 0.61 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.98 | 0.10 | 0.05 | 0.96 | 0.11 | 0.06 | 0.92 | 0.55 | 0.39 | 0.98 | 0.74 | 0.59 | 1.00 | 0.22 | 0.13 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| dog: weighted | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.77 | 0.68 | 0.61 | 0.97 | 0.75 | 0.60 | 0.86 | 0.71 | 0.61 | 0.92 | 0.13 | 0.07 | 0.91 | 0.14 | 0.07 | 0.93 | 0.55 | 0.39 | 0.96 | 0.71 | 0.57 | 1.00 | 0.32 | 0.20 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 1 | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.77 | 0.68 | 0.61 | 0.97 | 0.75 | 0.60 | 0.86 | 0.71 | 0.61 | 0.93 | 0.17 | 0.09 | 0.92 | 0.18 | 0.10 | 0.93 | 0.54 | 0.39 | 0.96 | 0.72 | 0.58 | 1.00 | 0.30 | 0.17 | 0.99 | 0.75 | 0.61 | NaN | NaN | 0.00 |
| 2 | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.77 | 0.68 | 0.61 | 0.97 | 0.75 | 0.60 | 0.86 | 0.71 | 0.61 | 0.91 | 0.09 | 0.05 | 0.90 | 0.09 | 0.05 | 0.93 | 0.55 | 0.39 | 0.96 | 0.71 | 0.56 | 1.00 | 0.41 | 0.26 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 3 | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.78 | 0.68 | 0.61 | 0.97 | 0.75 | 0.60 | 0.86 | 0.71 | 0.61 | 0.91 | 0.13 | 0.07 | 0.90 | 0.14 | 0.07 | 0.93 | 0.55 | 0.39 | 0.96 | 0.72 | 0.58 | 1.00 | 0.22 | 0.13 | 0.99 | 0.75 | 0.61 | NaN | NaN | 0.00 |
| 4 | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.77 | 0.68 | 0.60 | 0.97 | 0.74 | 0.60 | 0.86 | 0.71 | 0.61 | 0.92 | 0.19 | 0.10 | 0.91 | 0.19 | 0.11 | 0.93 | 0.55 | 0.39 | 0.96 | 0.70 | 0.55 | 1.00 | 0.45 | 0.29 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
| 5 | 1.00 | 1.00 | 1.00 | 0.74 | 0.60 | 0.50 | 0.77 | 0.68 | 0.61 | 0.97 | 0.75 | 0.60 | 0.86 | 0.71 | 0.61 | 0.91 | 0.09 | 0.05 | 0.90 | 0.10 | 0.05 | 0.93 | 0.55 | 0.39 | 0.96 | 0.72 | 0.58 | 1.00 | 0.22 | 0.13 | 0.99 | 0.75 | 0.60 | NaN | NaN | 0.00 |
n/a: result alignment not provided or not readable
NaN: division per zero, likely due to empty alignment.
This track has been performed by Jérôme Euzenat. If you have any problems working with the ontologies, any questions or suggestions, feel free to contact him
[1] Jérôme Euzenat, Maria Roşoiu, Cássia Trojahn dos Santos. Ontology matching benchmarks: generation, stability, and discriminability, Journal of web semantics 21:30-48, 2013 [DOI:10.1016/j.websem.2013.05.002]